Top-Down Proteomics and the Challenges of True Proteoform Characterization
- PMID: 37937372
- PMCID: PMC10696603
- DOI: 10.1021/acs.jproteome.3c00416
Top-Down Proteomics and the Challenges of True Proteoform Characterization
Abstract
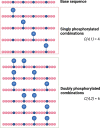
Top-down proteomics (TDP) aims to identify and profile intact protein forms (proteoforms) extracted from biological samples. True proteoform characterization requires that both the base protein sequence be defined and any mass shifts identified, ideally localizing their positions within the protein sequence. Being able to fully elucidate proteoform profiles lends insight into characterizing proteoform-unique roles, and is a crucial aspect of defining protein structure-function relationships and the specific roles of different (combinations of) protein modifications. However, defining and pinpointing protein post-translational modifications (PTMs) on intact proteins remains a challenge. Characterization of (heavily) modified proteins (>∼30 kDa) remains problematic, especially when they exist in a population of similarly modified, or kindred, proteoforms. This issue is compounded as the number of modifications increases, and thus the number of theoretical combinations. Here, we present our perspective on the challenges of analyzing kindred proteoform populations, focusing on annotation of protein modifications on an "average" protein. Furthermore, we discuss the technical requirements to obtain high quality fragmentation spectral data to robustly define site-specific PTMs, and the fact that this is tempered by the time requirements necessary to separate proteoforms in advance of mass spectrometry analysis.
Keywords: phosphorylation; post-translational modification; proteoform; top-down proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Characterization of Proteoforms with Unknown Post-translational Modifications Using the MIScore.J Proteome Res. 2016 Aug 5;15(8):2422-32. doi: 10.1021/acs.jproteome.5b01098. Epub 2016 Jul 1. J Proteome Res. 2016. PMID: 27291504 Free PMC article.
-
Expanding Proteoform Identifications in Top-Down Proteomic Analyses by Constructing Proteoform Families.Anal Chem. 2018 Jan 16;90(2):1325-1333. doi: 10.1021/acs.analchem.7b04221. Epub 2017 Dec 22. Anal Chem. 2018. PMID: 29227670 Free PMC article.
-
Improving Proteoform Identifications in Complex Systems Through Integration of Bottom-Up and Top-Down Data.J Proteome Res. 2020 Aug 7;19(8):3510-3517. doi: 10.1021/acs.jproteome.0c00332. Epub 2020 Jul 10. J Proteome Res. 2020. PMID: 32584579 Free PMC article.
-
Identification and Quantification of Proteoforms by Mass Spectrometry.Proteomics. 2019 May;19(10):e1800361. doi: 10.1002/pmic.201800361. Proteomics. 2019. PMID: 31050378 Free PMC article. Review.
-
Novel Strategies to Address the Challenges in Top-Down Proteomics.J Am Soc Mass Spectrom. 2021 Jun 2;32(6):1278-1294. doi: 10.1021/jasms.1c00099. Epub 2021 May 13. J Am Soc Mass Spectrom. 2021. PMID: 33983025 Free PMC article. Review.
Cited by
-
Top-down proteomics.Nat Rev Methods Primers. 2024;4(1):38. doi: 10.1038/s43586-024-00318-2. Epub 2024 Jun 13. Nat Rev Methods Primers. 2024. PMID: 39006170 Free PMC article.
-
I'm Walking into Spiderwebs: Making Sense of Protein-Protein Interaction Data.J Proteome Res. 2024 Aug 2;23(8):2723-2732. doi: 10.1021/acs.jproteome.3c00892. Epub 2024 Mar 31. J Proteome Res. 2024. PMID: 38556766 Free PMC article. Review.
-
Influence of different sample preparation approaches on proteoform identification by top-down proteomics.Nat Methods. 2024 Dec;21(12):2397-2407. doi: 10.1038/s41592-024-02481-6. Epub 2024 Oct 22. Nat Methods. 2024. PMID: 39438734 Free PMC article.
-
Considerations for defining +80 Da mass shifts in mass spectrometry-based proteomics: phosphorylation and beyond.Chem Commun (Camb). 2023 Sep 26;59(77):11484-11499. doi: 10.1039/d3cc02909c. Chem Commun (Camb). 2023. PMID: 37681662 Free PMC article. Review.
-
Inside the chase after those elusive proteoforms.Nat Methods. 2024 Feb;21(2):158-163. doi: 10.1038/s41592-024-02170-4. Nat Methods. 2024. PMID: 38308015 No abstract available.
References
-
- Wilkins M. R.; Sanchez J. C.; Gooley A. A.; Appel R. D.; Humphery-Smith I.; Hochstrasser D. F.; Williams K. L. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng. Rev. 1996, 13, 19–50. 10.1080/02648725.1996.10647923. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

