Genetic risk factors for severe and fatigue dominant long COVID and commonalities with ME/CFS identified by combinatorial analysis
- PMID: 37915075
- PMCID: PMC10621206
- DOI: 10.1186/s12967-023-04588-4
Genetic risk factors for severe and fatigue dominant long COVID and commonalities with ME/CFS identified by combinatorial analysis
Abstract
Background: Long COVID is a debilitating chronic condition that has affected over 100 million people globally. It is characterized by a diverse array of symptoms, including fatigue, cognitive dysfunction and respiratory problems. Studies have so far largely failed to identify genetic associations, the mechanisms behind the disease, or any common pathophysiology with other conditions such as myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) that present with similar symptoms.
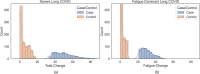
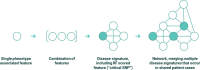
Methods: We used a combinatorial analysis approach to identify combinations of genetic variants significantly associated with the development of long COVID and to examine the biological mechanisms underpinning its various symptoms. We compared two subpopulations of long COVID patients from Sano Genetics' Long COVID GOLD study cohort, focusing on patients with severe or fatigue dominant phenotypes. We evaluated the genetic signatures previously identified in an ME/CFS population against this long COVID population to understand similarities with other fatigue disorders that may be triggered by a prior viral infection. Finally, we also compared the output of this long COVID analysis against known genetic associations in other chronic diseases, including a range of metabolic and neurological disorders, to understand the overlap of pathophysiological mechanisms.
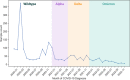
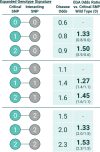
Results: Combinatorial analysis identified 73 genes that were highly associated with at least one of the long COVID populations included in this analysis. Of these, 9 genes have prior associations with acute COVID-19, and 14 were differentially expressed in a transcriptomic analysis of long COVID patients. A pathway enrichment analysis revealed that the biological pathways most significantly associated with the 73 long COVID genes were mainly aligned with neurological and cardiometabolic diseases. Expanded genotype analysis suggests that specific SNX9 genotypes are a significant contributor to the risk of or protection against severe long COVID infection, but that the gene-disease relationship is context dependent and mediated by interactions with KLF15 and RYR3. Comparison of the genes uniquely associated with the Severe and Fatigue Dominant long COVID patients revealed significant differences between the pathways enriched in each subgroup. The genes unique to Severe long COVID patients were associated with immune pathways such as myeloid differentiation and macrophage foam cells. Genes unique to the Fatigue Dominant subgroup were enriched in metabolic pathways such as MAPK/JNK signaling. We also identified overlap in the genes associated with Fatigue Dominant long COVID and ME/CFS, including several involved in circadian rhythm regulation and insulin regulation. Overall, 39 SNPs associated in this study with long COVID can be linked to 9 genes identified in a recent combinatorial analysis of ME/CFS patient from UK Biobank. Among the 73 genes associated with long COVID, 42 are potentially tractable for novel drug discovery approaches, with 13 of these already targeted by drugs in clinical development pipelines. From this analysis for example, we identified TLR4 antagonists as repurposing candidates with potential to protect against long term cognitive impairment pathology caused by SARS-CoV-2. We are currently evaluating the repurposing potential of these drug targets for use in treating long COVID and/or ME/CFS.
Conclusion: This study demonstrates the power of combinatorial analytics for stratifying heterogeneous populations in complex diseases that do not have simple monogenic etiologies. These results build upon the genetic findings from combinatorial analyses of severe acute COVID-19 patients and an ME/CFS population and we expect that access to additional independent, larger patient datasets will further improve the disease insights and validate potential treatment options in long COVID.
Keywords: Combinatorial analytics; Long COVID; ME/CFS; PASC; POTS; Patient stratification; Post-Covid; Post-acute COVID syndrome; Post-acute sequelae of COVID-19.
© 2023. The Author(s).
Conflict of interest statement
K.T., M.P., S.D., K.C., J.S, and S.G. are employees of PrecisionLife, Ltd. S.G. is a shareholder of PrecisionLife, Ltd.
Figures



Similar articles
-
Bioinformatics and systems biology approach to identify the pathogenetic link of Long COVID and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome.Front Immunol. 2022 Sep 16;13:952987. doi: 10.3389/fimmu.2022.952987. eCollection 2022. Front Immunol. 2022. PMID: 36189286 Free PMC article.
-
Cardiovascular and haematological pathology in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS): A role for viruses.Blood Rev. 2023 Jul;60:101075. doi: 10.1016/j.blre.2023.101075. Epub 2023 Mar 20. Blood Rev. 2023. PMID: 36963989 Free PMC article. Review.
-
Long-term neuromuscular consequences of SARS-Cov-2 and their similarities with myalgic encephalomyelitis/chronic fatigue syndrome: results of the retrospective CoLGEM study.J Transl Med. 2022 Sep 24;20(1):429. doi: 10.1186/s12967-022-03638-7. J Transl Med. 2022. PMID: 36153556 Free PMC article.
-
Orthostatic Symptoms and Reductions in Cerebral Blood Flow in Long-Haul COVID-19 Patients: Similarities with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome.Medicina (Kaunas). 2021 Dec 24;58(1):28. doi: 10.3390/medicina58010028. Medicina (Kaunas). 2021. PMID: 35056336 Free PMC article.
-
Analysis of post COVID-19 condition and its overlap with myalgic encephalomyelitis/chronic fatigue syndrome.J Adv Res. 2022 Sep;40:179-196. doi: 10.1016/j.jare.2021.11.013. Epub 2021 Nov 26. J Adv Res. 2022. PMID: 36100326 Free PMC article. Review.
Cited by
-
Mechanisms of long COVID and the path toward therapeutics.Cell. 2024 Oct 3;187(20):5500-5529. doi: 10.1016/j.cell.2024.07.054. Epub 2024 Sep 25. Cell. 2024. PMID: 39326415 Review.
-
Postacute Sequelae of COVID (PASC or Long COVID): An Evidenced-Based Approach.Open Forum Infect Dis. 2024 Aug 27;11(9):ofae462. doi: 10.1093/ofid/ofae462. eCollection 2024 Sep. Open Forum Infect Dis. 2024. PMID: 39220656 Free PMC article. Review.
-
Longitudinal cytokine and multi-modal health data of an extremely severe ME/CFS patient with HSD reveals insights into immunopathology, and disease severity.Front Immunol. 2024 Apr 8;15:1369295. doi: 10.3389/fimmu.2024.1369295. eCollection 2024. Front Immunol. 2024. PMID: 38650940 Free PMC article.
-
Host Genetic Variation Impacts SARS-CoV-2 Vaccination Response in the Diversity Outbred Mouse Population.Vaccines (Basel). 2024 Jan 20;12(1):103. doi: 10.3390/vaccines12010103. Vaccines (Basel). 2024. PMID: 38276675 Free PMC article.
-
Gut Microbiota and Mitochondria: Health and Pathophysiological Aspects of Long COVID.Int J Mol Sci. 2023 Dec 6;24(24):17198. doi: 10.3390/ijms242417198. Int J Mol Sci. 2023. PMID: 38139027 Free PMC article. Review.
References
-
- World Health Organization WHO Fact Sheets Post COVID-19 condition (Long COVID) available from https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-cond.... Accessed 8 Oct 2023
-
- O'Mahoney LL, Routen A, Gillies C, Ekezie W, Welford A, Zhang A, Karamchandani U, Simms-Williams N, Cassambai S, et al. The prevalence and long-term health effects of long Covid among hospitalised and non-hospitalised populations: a systematic review and meta-analysis. EClinicalMedicine. 2022;1(55):101762. doi: 10.1016/j.eclinm.2022.101762.Erratum.In:EClinicalMedicine.2023May;59:101959. - DOI - PMC - PubMed
-
- WHO Coronavirus (COVID-19) Dashboard https://covid19.who.int/, last Accessed 4 June 2023
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

