A carbon-nitrogen negative feedback loop underlies the repeated evolution of cnidarian-Symbiodiniaceae symbioses
- PMID: 37914686
- PMCID: PMC10620218
- DOI: 10.1038/s41467-023-42582-y
A carbon-nitrogen negative feedback loop underlies the repeated evolution of cnidarian-Symbiodiniaceae symbioses
Abstract
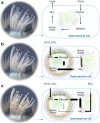
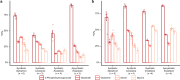
Symbiotic associations with Symbiodiniaceae have evolved independently across a diverse range of cnidarian taxa including reef-building corals, sea anemones, and jellyfish, yet the molecular mechanisms underlying their regulation and repeated evolution are still elusive. Here, we show that despite their independent evolution, cnidarian hosts use the same carbon-nitrogen negative feedback loop to control symbiont proliferation. Symbiont-derived photosynthates are used to assimilate nitrogenous waste via glutamine synthetase-glutamate synthase-mediated amino acid biosynthesis in a carbon-dependent manner, which regulates the availability of nitrogen to the symbionts. Using nutrient supplementation experiments, we show that the provision of additional carbohydrates significantly reduces symbiont density while ammonium promotes symbiont proliferation. High-resolution metabolic analysis confirmed that all hosts co-incorporated glucose-derived 13C and ammonium-derived 15N via glutamine synthetase-glutamate synthase-mediated amino acid biosynthesis. Our results reveal a general carbon-nitrogen negative feedback loop underlying these symbioses and provide a parsimonious explanation for their repeated evolution.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Coupled carbon and nitrogen cycling regulates the cnidarian-algal symbiosis.Nat Commun. 2023 Nov 1;14(1):6948. doi: 10.1038/s41467-023-42579-7. Nat Commun. 2023. PMID: 37914705 Free PMC article.
-
Symbiont population control by host-symbiont metabolic interaction in Symbiodiniaceae-cnidarian associations.Nat Commun. 2020 Jan 8;11(1):108. doi: 10.1038/s41467-019-13963-z. Nat Commun. 2020. PMID: 31913264 Free PMC article.
-
Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia.PLoS Genet. 2019 Jun 24;15(6):e1008189. doi: 10.1371/journal.pgen.1008189. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31233506 Free PMC article.
-
Innate immunity and cnidarian-Symbiodiniaceae mutualism.Dev Comp Immunol. 2019 Jan;90:199-209. doi: 10.1016/j.dci.2018.09.020. Epub 2018 Sep 27. Dev Comp Immunol. 2019. PMID: 30268783 Review.
-
How does an animal behave like a plant? Physiological and molecular adaptations of zooxanthellae and their hosts to symbiosis.C R Biol. 2018 May-Jun;341(5):276-280. doi: 10.1016/j.crvi.2018.03.007. Epub 2018 Apr 9. C R Biol. 2018. PMID: 29650460 Review.
Cited by
-
Using clusterProfiler to characterize multiomics data.Nat Protoc. 2024 Nov;19(11):3292-3320. doi: 10.1038/s41596-024-01020-z. Epub 2024 Jul 17. Nat Protoc. 2024. PMID: 39019974 Review.
-
Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana.mBio. 2024 Sep 11;15(9):e0105924. doi: 10.1128/mbio.01059-24. Epub 2024 Aug 6. mBio. 2024. PMID: 39105583 Free PMC article.
-
Coral larvae increase nitrogen assimilation to stabilize algal symbiosis and combat bleaching under increased temperature.PLoS Biol. 2024 Nov 12;22(11):e3002875. doi: 10.1371/journal.pbio.3002875. eCollection 2024 Nov. PLoS Biol. 2024. PMID: 39531470 Free PMC article.
-
Coupled carbon and nitrogen cycling regulates the cnidarian-algal symbiosis.Nat Commun. 2023 Nov 1;14(1):6948. doi: 10.1038/s41467-023-42579-7. Nat Commun. 2023. PMID: 37914705 Free PMC article.
-
Symbiodiniaceae algal symbionts of Pocillopora damicornis larvae provide more carbon to their coral host under elevated levels of acidification and temperature.Commun Biol. 2024 Nov 18;7(1):1528. doi: 10.1038/s42003-024-07203-4. Commun Biol. 2024. PMID: 39558079 Free PMC article.
References
-
- Stat M, Carter D, Hoegh-Guldberg O. The evolutionary history of Symbiodinium and scleractinian hosts—symbiosis, diversity, and the effect of climate change. Perspect. Plant Ecol. Evol. 2006;8:23–43. doi: 10.1016/j.ppees.2006.04.001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

