Identification and Characterization of a Novel Hypovirus from the Phytopathogenic Fungus Botryosphaeria dothidea
- PMID: 37896836
- PMCID: PMC10611357
- DOI: 10.3390/v15102059
Identification and Characterization of a Novel Hypovirus from the Phytopathogenic Fungus Botryosphaeria dothidea
Abstract
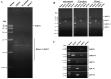
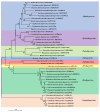
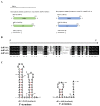
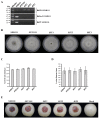
Many mycoviruses have been accurately and successfully identified in plant pathogenic fungus Botryosphaeria dothidea. This study discovered three mycoviruses from a B. dothidea strain SXD111 using high-throughput sequencing technology. A novel hypovirus was tentatively named Botryosphaeria dothidea hypovirus 1 (BdHV1/SXD111). The other two were known viruses, which we named Botryosphaeria dothidea polymycovirus 1 strain SXD111 (BdPmV1/SXD111) and Botryosphaeria dothidea partitivirus 1 strain SXD111 (BdPV1/SXD111). The genome of BdHV1/SXD111 is 11,128 nucleotides long, excluding the poly (A) tail. A papain-like cysteine protease (Pro), a UDP-glucose/sterol glucosyltransferase (UGT), an RNA-dependent RNA polyprotein (RdRp), and a helicase (Hel) were detected in the polyprotein of BdHV1/SXD111. Phylogenetic analysis showed that BdHV1/SXD111 was clustered with betahypovirus and separated from members of the other genera in the family Hypoviridae. The BdPmV1/SXD111 genome comprised five dsRNA segments with 2396, 2232, 1967, 1131, and 1060 bp lengths. Additionally, BdPV1/SXD111 harbored three dsRNA segments with 1823, 1623, and 557 bp lengths. Furthermore, the smallest dsRNA was a novel satellite component of BdPV1/SXD111. BdHV1/SXD111 could be transmitted through conidia and hyphae contact, whereas it likely has no apparent impact on the morphologies and virulence of the host fungus. Thus, this study is the first report of a betahypovirus isolated from the fungus B. dothidea. Importantly, our results significantly enhance the diversity of the B. dothidea viruses.
Keywords: Betahypovirus; Botryosphaeria dothidea; Botryosphaeria dothidea hypovirus 1; Hypoviridae; genome.
Conflict of interest statement
The authors declare no conflict of interes in this paper.
Figures

Similar articles
-
Hypovirulence of the phytopathogenic fungus Botryosphaeria dothidea: association with a coinfecting chrysovirus and a partitivirus.J Virol. 2014 Jul;88(13):7517-27. doi: 10.1128/JVI.00538-14. Epub 2014 Apr 23. J Virol. 2014. PMID: 24760881 Free PMC article.
-
Molecular characterization of a novel victorivirus isolated from Botryosphaeria dothidea, the causal agent of longan leaf spot disease.Arch Virol. 2022 Nov;167(11):2417-2422. doi: 10.1007/s00705-022-05573-w. Epub 2022 Aug 13. Arch Virol. 2022. PMID: 35962824
-
Characterization of a Novel Mycovirus from the Phytopathogenic Fungus Botryosphaeria dothidea.Viruses. 2022 Feb 6;14(2):331. doi: 10.3390/v14020331. Viruses. 2022. PMID: 35215923 Free PMC article.
-
Characterization of a Botybirnavirus Conferring Hypovirulence in the Phytopathogenic Fungus Botryosphaeria dothidea.Viruses. 2019 Mar 17;11(3):266. doi: 10.3390/v11030266. Viruses. 2019. PMID: 30884907 Free PMC article.
-
Botryosphaeria dothidea: a latent pathogen of global importance to woody plant health.Mol Plant Pathol. 2017 May;18(4):477-488. doi: 10.1111/mpp.12495. Epub 2016 Dec 13. Mol Plant Pathol. 2017. PMID: 27682468 Free PMC article. Review.
References
-
- Ayllón M.A., Vainio E.J. Mycoviruses as a part of the global virome: Diversity, evolutionary links and lifestyle. Adv. Virus Res. 2023;115:1–86. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

