Polyploid Giant Cancer Cells Generated from Human Cytomegalovirus-Infected Prostate Epithelial Cells
- PMID: 37894361
- PMCID: PMC10604969
- DOI: 10.3390/cancers15204994
Polyploid Giant Cancer Cells Generated from Human Cytomegalovirus-Infected Prostate Epithelial Cells
Abstract
Background: Prostate cancer is the most commonly diagnosed malignancy and the sixth leading cause of cancer death in men worldwide. Chromosomal instability (CIN) and polyploid giant cancer cells (PGCCs) have been considered predominant hallmarks of cancer. Recent clinical studies have proven the association of CIN, aneuploidy, and PGCCs with poor prognosis of prostate cancer (PCa). Evidence of HCMV transforming potential might indicate that HCMV may be involved in PCa.
Methods: Herein, we underline the role of the high-risk HCMV-DB and -BL clinical strains in transforming prostate epithelial cells and assess the molecular and cellular oncogenic processes associated with PCa.
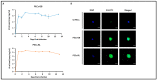
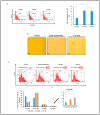
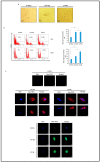
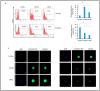
Results: Oncogenesis parallels a sustained growth of "CMV-Transformed Prostate epithelial cells" or CTP cells that highly express Myc and EZH2, forming soft agar colonies and displaying stemness as well as mesenchymal features, hence promoting EMT as well as PGCCs and a spheroid appearance.
Conclusions: HCMV-induced Myc and EZH2 upregulation coupled with stemness and EMT traits in IE1-expressing CTP might highlight the potential role of HCMV in PCa development and encourage the use of anti-EZH2 and anti-HCMV in PCa treatment.
Keywords: CIN; PGCCs; high-risk HCMV strains; human cytomegalovirus; oncogenesis; prostate cancer; stemness.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Polyploid Giant Cancer Cells: A Distinctive Feature in the Transformation of Epithelial Cells by High-Risk Oncogenic HCMV Strains.Viruses. 2024 Jul 31;16(8):1225. doi: 10.3390/v16081225. Viruses. 2024. PMID: 39205199 Free PMC article. Review.
-
Polyploid giant cancer cells, EZH2 and Myc upregulation in mammary epithelial cells infected with high-risk human cytomegalovirus.EBioMedicine. 2022 Jun;80:104056. doi: 10.1016/j.ebiom.2022.104056. Epub 2022 May 18. EBioMedicine. 2022. PMID: 35596973 Free PMC article.
-
Oncogenic and Stemness Signatures of the High-Risk HCMV Strains in Breast Cancer Progression.Cancers (Basel). 2022 Sep 1;14(17):4271. doi: 10.3390/cancers14174271. Cancers (Basel). 2022. PMID: 36077806 Free PMC article.
-
Polyploid giant cancer cells, cytokines and cytomegalovirus in breast cancer progression.Cancer Cell Int. 2023 Jun 20;23(1):119. doi: 10.1186/s12935-023-02971-1. Cancer Cell Int. 2023. PMID: 37340387 Free PMC article.
-
High-Risk Oncogenic Human Cytomegalovirus.Viruses. 2022 Nov 7;14(11):2462. doi: 10.3390/v14112462. Viruses. 2022. PMID: 36366560 Free PMC article. Review.
Cited by
-
Cellular Transformation by Human Cytomegalovirus.Cancers (Basel). 2024 May 22;16(11):1970. doi: 10.3390/cancers16111970. Cancers (Basel). 2024. PMID: 38893091 Free PMC article.
-
The Role of Oncogenic Viruses in the Pathogenesis of Sporadic Breast Cancer: A Comprehensive Review of the Current Literature.Pathogens. 2024 May 25;13(6):451. doi: 10.3390/pathogens13060451. Pathogens. 2024. PMID: 38921749 Free PMC article. Review.
-
Polyploidy in Cancer: Causal Mechanisms, Cancer-Specific Consequences, and Emerging Treatments.Mol Cancer Ther. 2024 May 2;23(5):638-647. doi: 10.1158/1535-7163.MCT-23-0578. Mol Cancer Ther. 2024. PMID: 38315992 Free PMC article. Review.
-
Polyploid Giant Cancer Cells: A Distinctive Feature in the Transformation of Epithelial Cells by High-Risk Oncogenic HCMV Strains.Viruses. 2024 Jul 31;16(8):1225. doi: 10.3390/v16081225. Viruses. 2024. PMID: 39205199 Free PMC article. Review.
-
Generation of glioblastoma in mice engrafted with human cytomegalovirus-infected astrocytes.Cancer Gene Ther. 2024 Jul;31(7):1070-1080. doi: 10.1038/s41417-024-00767-7. Epub 2024 Mar 29. Cancer Gene Ther. 2024. PMID: 38553638 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

