Regulation of self-renewal and senescence in primitive mesenchymal stem cells by Wnt and TGFβ signaling
- PMID: 37880755
- PMCID: PMC10601332
- DOI: 10.1186/s13287-023-03533-y
Regulation of self-renewal and senescence in primitive mesenchymal stem cells by Wnt and TGFβ signaling
Abstract
Background: The therapeutic application of multipotent mesenchymal stem cells (MSCs) encounters significant challenges, primarily stemming from their inadequate growth and limited self-renewal capabilities. Additionally, as MSCs are propagated, their ability to self-renew declines, and the exact cellular and molecular changes responsible for this are poorly understood. This study aims to uncover the complex molecular mechanisms that govern the self-renewal of primitive (p) MSCs.
Methods: We grew pMSCs using two types of medium, fetal bovine serum (FM) and xeno-free (XM), at both low passage (LP, P3) and high passage (HP, P20). To evaluate LP and HP pMSCs, we examined their physical characteristics, cell surface markers, growth rate, colony-forming ability, BrdU assays for proliferation, telomerase activity, and potential to differentiate into three lineages. Moreover, we conducted RNA-seq to analyze their transcriptome and MNase-seq analysis to investigate nucleosome occupancies.
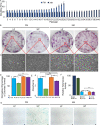
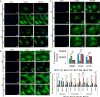
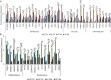
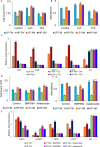
Results: When grown in FM, pMSCs underwent changes in their cellular morphology, becoming larger and elongated. This was accompanied by a decrease in the expression of CD90 and CD49f, as well as a reduction in CFE, proliferation rate, and telomerase activity. In addition, these cells showed an increased tendency to differentiate into the adipogenic lineage. However, when grown in XM, pMSCs maintained their self-renewal capacity and ability to differentiate into multiple lineages while preserving their fibroblastoid morphology. Transcriptomic analysis showed an upregulation of genes associated with self-renewal, cell cycle regulation, and DNA replication in XM-cultured pMSCs, while senescence-related genes were upregulated in FM-cultured cells. Further analysis demonstrated differential nucleosomal occupancies in self-renewal and senescence-related genes for pMSCs grown in XM and FM, respectively. These findings were confirmed by qRT-PCR analysis, which revealed alterations in the expression of genes related to self-renewal, cell cycle regulation, DNA replication, differentiation, and senescence. To understand the underlying mechanisms, we investigated the involvement of Wnt and TGFβ signaling pathways by modulating them with agonists and antagonists. This experimental manipulation led to the upregulation and downregulation of self-renewal genes in pMSCs, providing further insights into the signaling pathways governing the self-renewal and senescence of pMSCs.
Conclusion: Our study shows that the self-renewal potential of pMSCs is associated with the Wnt pathway, while senescence is linked to TGFβ.
Keywords: Differentiation; Mesenchymal stem cells; Proliferation; Self-renewal; Senescence; TGFβ signaling; Wnt pathway.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Update of
-
WNT and VEGF/PDGF signaling regulate self-renewal in primitive mesenchymal stem cells.Res Sq [Preprint]. 2023 Apr 10:rs.3.rs-2512048. doi: 10.21203/rs.3.rs-2512048/v1. Res Sq. 2023. Update in: Stem Cell Res Ther. 2023 Oct 26;14(1):305. doi: 10.1186/s13287-023-03533-y PMID: 37090660 Free PMC article. Updated. Preprint.
Similar articles
-
WNT and VEGF/PDGF signaling regulate self-renewal in primitive mesenchymal stem cells.Res Sq [Preprint]. 2023 Apr 10:rs.3.rs-2512048. doi: 10.21203/rs.3.rs-2512048/v1. Res Sq. 2023. Update in: Stem Cell Res Ther. 2023 Oct 26;14(1):305. doi: 10.1186/s13287-023-03533-y PMID: 37090660 Free PMC article. Updated. Preprint.
-
TZAP plays an inhibitory role in the self-renewal of porcine mesenchymal stromal cells and is implicated the regulation of premature senescence via the p53 pathway.J Transl Med. 2019 Mar 7;17(1):72. doi: 10.1186/s12967-019-1820-8. J Transl Med. 2019. PMID: 30845965 Free PMC article.
-
Low-dose telomerase is required for the expansion and migration of placental mesenchymal stem cells.Biochem Biophys Res Commun. 2022 Dec 25;636(Pt 2):40-47. doi: 10.1016/j.bbrc.2022.10.093. Epub 2022 Oct 31. Biochem Biophys Res Commun. 2022. PMID: 36343489
-
[DEVELOPMENT PROGRESS OF MESENCHYMAL STEM CELLS SENESCENCE].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2015 Jul;29(7):899-904. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2015. PMID: 26540988 Review. Chinese.
-
Mesenchymal stem cells derived from hard palate: An attractive alternative for regenerative medicine.Oral Dis. 2024 Oct;30(7):4145-4151. doi: 10.1111/odi.15043. Epub 2024 Jul 15. Oral Dis. 2024. PMID: 39007203 Review.
Cited by
-
Comparative Analysis of Serum and Serum-Free Medium Cultured Mesenchymal Stromal Cells for Cartilage Repair.Int J Mol Sci. 2024 Oct 2;25(19):10627. doi: 10.3390/ijms251910627. Int J Mol Sci. 2024. PMID: 39408956 Free PMC article.
-
Super-enhancer omics in stem cell.Mol Cancer. 2024 Aug 1;23(1):153. doi: 10.1186/s12943-024-02066-z. Mol Cancer. 2024. PMID: 39090713 Free PMC article. Review.
-
Potential Mechanism and Perspectives of Mesenchymal Stem Cell Therapy for Ischemic Stroke: A Review.Glob Med Genet. 2024 Sep 2;11(4):278-284. doi: 10.1055/s-0044-1790231. eCollection 2024 Dec. Glob Med Genet. 2024. PMID: 39224463 Free PMC article. Review.
-
The Myofibroblast Fate of Therapeutic Mesenchymal Stromal Cells: Regeneration, Repair, or Despair?Int J Mol Sci. 2024 Aug 9;25(16):8712. doi: 10.3390/ijms25168712. Int J Mol Sci. 2024. PMID: 39201399 Free PMC article. Review.
-
Unlocking the versatile potential: Adipose-derived mesenchymal stem cells in ocular surface reconstruction and oculoplastics.World J Stem Cells. 2024 Feb 26;16(2):89-101. doi: 10.4252/wjsc.v16.i2.89. World J Stem Cells. 2024. PMID: 38455097 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

