This is a preprint.
A PARP14/TARG1-Regulated RACK1 MARylation Cycle Drives Stress Granule Dynamics in Ovarian Cancer Cells
- PMID: 37873085
- PMCID: PMC10592810
- DOI: 10.1101/2023.10.13.562273
A PARP14/TARG1-Regulated RACK1 MARylation Cycle Drives Stress Granule Dynamics in Ovarian Cancer Cells
Abstract
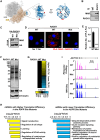
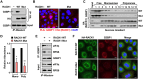
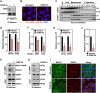
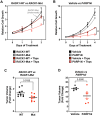
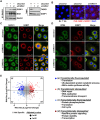
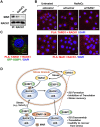
Mono(ADP-ribosyl)ation (MARylation) is emerging as a critical regulator of ribosome function and translation. Herein, we demonstrate that RACK1, an integral component of the ribosome, is MARylated on three acidic residues by the mono(ADP-ribosyl) transferase (MART) PARP14 in ovarian cancer cells. MARylation of RACK1 is required for stress granule formation and promotes the colocalization of RACK1 in stress granules with G3BP1, eIF3η, and 40S ribosomal proteins. In parallel, we observed reduced translation of a subset of mRNAs, including those encoding key cancer regulators (e.g., AKT). Treatment with a PARP14 inhibitor or mutation of the sites of MARylation on RACK1 blocks these outcomes, as well as the growth of ovarian cancer cells in culture and in vivo. To re-set the system after prolonged stress and recovery, the ADP-ribosyl hydrolase TARG1 deMARylates RACK1, leading to the dissociation of the stress granules and the restoration of translation. Collectively, our results demonstrate a therapeutically targetable pathway that controls stress granule assembly and disassembly in ovarian cancer cells.
Conflict of interest statement
Conflict of Interest Statement: W.L.K. is a founder, member of the SAB, member of the BOD, and a stockholder for ARase Therapeutics, Inc. He is also coholder of U.S. Patent 9,599,606 covering the ADP-ribose detection reagents used herein, which has been licensed to and is sold by EMD Millipore.
Figures
Similar articles
-
RACK1 MARylation regulates translation and stress granules in ovarian cancer cells.J Cell Biol. 2025 Feb 3;224(2):e202401101. doi: 10.1083/jcb.202401101. Epub 2025 Jan 6. J Cell Biol. 2025. PMID: 39760726 Free PMC article.
-
Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers.Cell. 2021 Aug 19;184(17):4531-4546.e26. doi: 10.1016/j.cell.2021.07.005. Epub 2021 Jul 26. Cell. 2021. PMID: 34314702 Free PMC article.
-
MARTs and MARylation in the Cytosol: Biological Functions, Mechanisms of Action, and Therapeutic Potential.Cells. 2021 Feb 3;10(2):313. doi: 10.3390/cells10020313. Cells. 2021. PMID: 33546365 Free PMC article. Review.
-
The Controversial Roles of ADP-Ribosyl Hydrolases MACROD1, MACROD2 and TARG1 in Carcinogenesis.Cancers (Basel). 2020 Mar 5;12(3):604. doi: 10.3390/cancers12030604. Cancers (Basel). 2020. PMID: 32151005 Free PMC article. Review.
-
RBN-2397, a PARP7 Inhibitor, Synergizes with Paclitaxel to Inhibit Proliferation and Migration of Ovarian Cancer Cells.bioRxiv [Preprint]. 2024 Sep 5:2024.08.20.608802. doi: 10.1101/2024.08.20.608802. bioRxiv. 2024. PMID: 39229139 Free PMC article. Preprint.
References
-
- Ahmed S., Bott D., Gomez A., Tamblyn L., Rasheed A., Cho T., MacPherson L., Sugamori K.S., Yang Y., Grant D.M., Cummins C.L., and Matthews J.. 2015. Loss of the mono-ADP-ribosyltransferase, Tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J Biol Chem. 290:16824–16840. - PMC - PubMed
-
- Andrews S. 2010. FASTQC. A quality control tool for high throughput sequence data.
-
- Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., and Takekawa M.. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nature cell biology. 10:1324–1332. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
