The Many Faces of Histidine Triad Nucleotide Binding Protein 1 (HINT1)
- PMID: 37854629
- PMCID: PMC10580397
- DOI: 10.1021/acsptsci.3c00079
The Many Faces of Histidine Triad Nucleotide Binding Protein 1 (HINT1)
Abstract
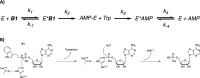
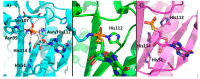
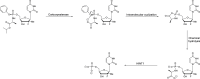
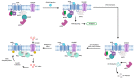
The histidine triad nucleotide binding protein 1 (HINT1) is a nucleoside phosphoramidase that has garnered interest due to its widespread expression and participation in a broad range of biological processes. Herein, we discuss the role of HINT1 as a regulator of several CNS functions, tumor suppressor, and mast cell activator via its interactions with multiple G-protein-coupled receptors and transcription factors. Importantly, altered HINT1 expression and mutation are connected to the progression of multiple disease states, including several neuropsychiatric disorders, peripheral neuropathy, and tumorigenesis. Additionally, due to its involvement in the activation of several clinically used phosphoramidate prodrugs, tremendous efforts have been made to better understand the interactions behind nucleoside binding and phosphoramidate hydrolysis by HINT1. We detail the substrate specificity and catalytic mechanism of HINT1 hydrolysis, while highlighting the structural biology behind these efforts. The aim of this review is to summarize the multitude of biological and pharmacological functions in which HINT1 participates while addressing the areas of need for future research.
© 2023 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

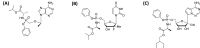
Similar articles
-
Evidence that human histidine triad nucleotide binding protein 3 (Hint3) is a distinct branch of the histidine triad (HIT) superfamily.J Mol Biol. 2007 Nov 2;373(4):978-89. doi: 10.1016/j.jmb.2007.08.023. Epub 2007 Aug 21. J Mol Biol. 2007. PMID: 17870088
-
Phosphoramidate pronucleotides: a comparison of the phosphoramidase substrate specificity of human and Escherichia coli histidine triad nucleotide binding proteins.Mol Pharm. 2007 Mar-Apr;4(2):208-17. doi: 10.1021/mp060070y. Epub 2007 Jan 12. Mol Pharm. 2007. PMID: 17217311
-
Emerging Roles of Histidine Triad Nucleotide Binding Protein 1 in Neuropsychiatric Diseases.Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2017 Oct 30;39(5):705-714. doi: 10.3881/j.issn.1000-503X.2017.05.018. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2017. PMID: 29125116 Review. English.
-
The oncogenic and immunological roles of histidine triad nucleotide-binding protein 1 in human cancers and their experimental validation in the MCF-7 cell line.Ann Transl Med. 2023 Feb 15;11(3):147. doi: 10.21037/atm-22-6637. Ann Transl Med. 2023. PMID: 36846002 Free PMC article.
-
[HINT1--a novel tumor suppressor protein of the HIT superfamily].Postepy Biochem. 2010;56(1):55-60. Postepy Biochem. 2010. PMID: 20499681 Review. Polish.
Cited by
-
HINT1 Gene Polymorphisms, Smoking Behaviour, and Personality Traits: A Haplotype Case-Control Study.Int J Mol Sci. 2024 Jul 12;25(14):7657. doi: 10.3390/ijms25147657. Int J Mol Sci. 2024. PMID: 39062900 Free PMC article.
References
-
- Krakowiak A.; Fryc I. Histidine triad protein superfamily-biological function and enzymatic activity. Postepy Biochem. 2012, 58 (3), 302–313. - PubMed
-
(in Polish).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
