This is a preprint.
Microglial over-pruning of synapses during development in autism-associated SCN2A-deficient mice and human cerebral organoids
- PMID: 37841865
- PMCID: PMC10571631
- DOI: 10.21203/rs.3.rs-3270664/v1
Microglial over-pruning of synapses during development in autism-associated SCN2A-deficient mice and human cerebral organoids
Update in
-
Microglial over-pruning of synapses during development in autism-associated SCN2A-deficient mice and human cerebral organoids.Mol Psychiatry. 2024 Aug;29(8):2424-2437. doi: 10.1038/s41380-024-02518-4. Epub 2024 Mar 18. Mol Psychiatry. 2024. PMID: 38499656
Abstract
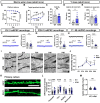
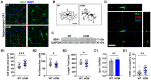
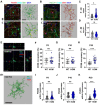
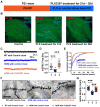
Autism spectrum disorder (ASD) is a major neurodevelopmental disorder affecting 1 in 36 children in the United States. While neurons have been the focus to understand ASD, an altered neuro-immune response in the brain may be closely associated with ASD, and a neuro-immune interaction could play a role in the disease progression. As the resident immune cells of the brain, microglia regulate brain development and homeostasis via core functions including phagocytosis of synapses. While ASD has been traditionally considered a polygenic disorder, recent large-scale human genetic studies have identified SCN2A deficiency as a leading monogenic cause of ASD and intellectual disability. We generated a Scn2a-deficient mouse model, which displays major behavioral and neuronal phenotypes. However, the role of microglia in this disease model is unknown. Here, we reported that Scn2a-deficient mice have impaired learning and memory, accompanied by reduced synaptic transmission and lower spine density in neurons of the hippocampus. Microglia in Scn2a-deficient mice are partially activated, exerting excessive phagocytic pruning of post-synapses related to the complement C3 cascades during selective developmental stages. The ablation of microglia using PLX3397 partially restores synaptic transmission and spine density. To extend our findings from rodents to human cells, we established a microglial-incorporated human cerebral organoid model carrying an SCN2A protein-truncating mutation identified in children with ASD. We found that human microglia display increased elimination of post-synapse in cerebral organoids carrying the SCN2A mutation. Our study establishes a key role of microglia in multi-species autism-associated models of SCN2A deficiency from mouse to human cells.
Keywords: Development; Microglia; Organoids; SCN2A; Synaptic pruning.
Conflict of interest statement
COMPETING INTERESTS The authors declare no conflict of interest.
Figures

Similar articles
-
Microglial over-pruning of synapses during development in autism-associated SCN2A-deficient mice and human cerebral organoids.Mol Psychiatry. 2024 Aug;29(8):2424-2437. doi: 10.1038/s41380-024-02518-4. Epub 2024 Mar 18. Mol Psychiatry. 2024. PMID: 38499656
-
Microglial Tmem59 Deficiency Impairs Phagocytosis of Synapse and Leads to Autism-Like Behaviors in Mice.J Neurosci. 2022 Jun 22;42(25):4958-4979. doi: 10.1523/JNEUROSCI.1644-21.2022. Epub 2022 May 23. J Neurosci. 2022. PMID: 35606143 Free PMC article.
-
Microglia in the pathogenesis of autism spectrum disorders.Neurosci Res. 2015 Nov;100:1-5. doi: 10.1016/j.neures.2015.06.005. Epub 2015 Jun 25. Neurosci Res. 2015. PMID: 26116891 Review.
-
P2Y6 Receptor-Dependent Microglial Phagocytosis of Synapses during Development Regulates Synapse Density and Memory.J Neurosci. 2023 Nov 29;43(48):8090-8103. doi: 10.1523/JNEUROSCI.1089-23.2023. J Neurosci. 2023. PMID: 37758475 Free PMC article.
-
Microglia: Synaptic modulator in autism spectrum disorder.Front Psychiatry. 2022 Nov 17;13:958661. doi: 10.3389/fpsyt.2022.958661. eCollection 2022. Front Psychiatry. 2022. PMID: 36465285 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

