Establishment of a transparent soil system to study Bacillus subtilis chemical ecology
- PMID: 37838789
- PMCID: PMC10576751
- DOI: 10.1038/s43705-023-00318-5
Establishment of a transparent soil system to study Bacillus subtilis chemical ecology
Abstract
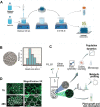
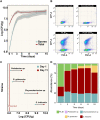
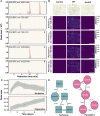
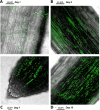
Bacterial secondary metabolites are structurally diverse molecules that drive microbial interaction by altering growth, cell differentiation, and signaling. Bacillus subtilis, a Gram-positive soil-dwelling bacterium, produces a wealth of secondary metabolites, among them, lipopeptides have been vastly studied by their antimicrobial, antitumor, and surfactant activities. However, the natural functions of secondary metabolites in the lifestyles of the producing organism remain less explored under natural conditions, i.e. in soil. Here, we describe a hydrogel-based transparent soil system to investigate B. subtilis chemical ecology under controllable soil-like conditions. The transparent soil matrix allows the growth of B. subtilis and other isolates gnotobiotically and under nutrient-controlled conditions. Additionally, we show that transparent soil allows the detection of lipopeptides production and dynamics by HPLC-MS, and MALDI-MS imaging, along with fluorescence imaging of 3-dimensional bacterial assemblages. We anticipate that this affordable and highly controllable system will promote bacterial chemical ecology research and help to elucidate microbial interactions driven by secondary metabolites.
© 2023. ISME Publications B.V.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Genomic and Chemical Diversity of Bacillus subtilis Secondary Metabolites against Plant Pathogenic Fungi.mSystems. 2021 Feb 23;6(1):e00770-20. doi: 10.1128/mSystems.00770-20. mSystems. 2021. PMID: 33622852 Free PMC article.
-
Lipopeptide Interplay Mediates Molecular Interactions between Soil Bacilli and Pseudomonads.Microbiol Spectr. 2021 Dec 22;9(3):e0203821. doi: 10.1128/spectrum.02038-21. Epub 2021 Dec 8. Microbiol Spectr. 2021. PMID: 34878336 Free PMC article.
-
Secondary metabolites of Bacillus subtilis impact the assembly of soil-derived semisynthetic bacterial communities.Beilstein J Org Chem. 2020 Dec 4;16:2983-2998. doi: 10.3762/bjoc.16.248. eCollection 2020. Beilstein J Org Chem. 2020. PMID: 33335606 Free PMC article.
-
Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics.FEMS Microbiol Rev. 2010 Nov;34(6):1037-62. doi: 10.1111/j.1574-6976.2010.00221.x. FEMS Microbiol Rev. 2010. PMID: 20412310 Review.
-
Bioactive Secondary Metabolites from Bacillus subtilis: A Comprehensive Review.J Nat Prod. 2019 Jul 26;82(7):2038-2053. doi: 10.1021/acs.jnatprod.9b00110. Epub 2019 Jul 9. J Nat Prod. 2019. PMID: 31287310 Review.
Cited by
-
Resistance towards and biotransformation of a Pseudomonas-produced secondary metabolite during community invasion.ISME J. 2024 Jan 8;18(1):wrae105. doi: 10.1093/ismejo/wrae105. ISME J. 2024. PMID: 38874164 Free PMC article.
-
Construction and characterisation of a structured, tuneable, and transparent 3D culture platform for soil bacteria.Microbiology (Reading). 2024 Jan;170(1):001429. doi: 10.1099/mic.0.001429. Microbiology (Reading). 2024. PMID: 38289644 Free PMC article.
-
Microbe-cellulose hydrogels as a model system for particulate carbon degradation in soil aggregates.ISME Commun. 2024 May 4;4(1):ycae068. doi: 10.1093/ismeco/ycae068. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38800124 Free PMC article.
-
Complex extracellular biology drives surface competition during colony expansion in Bacillus subtilis.ISME J. 2022 Oct;16(10):2320-2328. doi: 10.1038/s41396-022-01279-8. Epub 2022 Jul 5. ISME J. 2022. PMID: 35790818 Free PMC article.
References
-
- Saleem M, Hu J, Jousset A. More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health. Annu Rev Ecol Evol Syst. 2019;50:145–68. doi: 10.1146/annurev-ecolsys-110617-062605. - DOI
Grants and funding
- NNF19OC0055625/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF19SA0059360/Novo Nordisk Fonden (Novo Nordisk Foundation)
- DNRF137/Danmarks Grundforskningsfond (Danish National Research Foundation)
- DNRF137/Danmarks Grundforskningsfond (Danish National Research Foundation)
- DNRF137/Danmarks Grundforskningsfond (Danish National Research Foundation)

