Antiviral responses are shaped by heterogeneity in viral replication dynamics
- PMID: 37814072
- PMCID: PMC10627821
- DOI: 10.1038/s41564-023-01501-z
Antiviral responses are shaped by heterogeneity in viral replication dynamics
Abstract
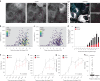
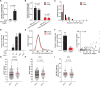
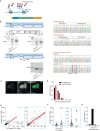
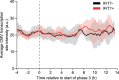
Antiviral signalling, which can be activated in host cells upon virus infection, restricts virus replication and communicates infection status to neighbouring cells. The antiviral response is heterogeneous, both quantitatively (efficiency of response activation) and qualitatively (transcribed antiviral gene set). To investigate the basis of this heterogeneity, we combined Virus Infection Real-time IMaging (VIRIM), a live-cell single-molecule imaging method, with real-time readouts of the dsRNA sensing pathway to analyse the response of human cells to encephalomyocarditis virus (EMCV) infection. We find that cell-to-cell heterogeneity in viral replication rates early in infection affect the efficiency of antiviral response activation, with lower replication rates leading to more antiviral response activation. Furthermore, we show that qualitatively distinct antiviral responses can be linked to the strength of the antiviral signalling pathway. Our analyses identify variation in early viral replication rates as an important parameter contributing to heterogeneity in antiviral response activation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
Encephalomyocarditis Virus Abrogates the Interferon Beta Signaling Pathway via Its Structural Protein VP2.J Virol. 2021 Feb 24;95(6):e01590-20. doi: 10.1128/JVI.01590-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33328314 Free PMC article.
-
Exploitation of nuclear protein SFPQ by the encephalomyocarditis virus to facilitate its replication.Biochem Biophys Res Commun. 2019 Feb 26;510(1):65-71. doi: 10.1016/j.bbrc.2019.01.032. Epub 2019 Jan 18. Biochem Biophys Res Commun. 2019. PMID: 30661786
-
Cholesterol 25-hydroxylase inhibits encephalomyocarditis virus replication through enzyme activity-dependent and independent mechanisms.Vet Microbiol. 2020 Jun;245:108658. doi: 10.1016/j.vetmic.2020.108658. Epub 2020 Apr 10. Vet Microbiol. 2020. PMID: 32456829
-
Macrophage Expression of Inflammatory Genes in Response to EMCV Infection.Biomolecules. 2015 Aug 18;5(3):1938-54. doi: 10.3390/biom5031938. Biomolecules. 2015. PMID: 26295266 Free PMC article. Review.
-
[Influenza viruses and intracellular signalling pathways].Berl Munch Tierarztl Wochenschr. 2006 Mar-Apr;119(3-4):101-11. Berl Munch Tierarztl Wochenschr. 2006. PMID: 16573200 Review. German.
Cited by
-
Boosting the toolbox for live imaging of translation.RNA. 2024 Sep 16;30(10):1374-1394. doi: 10.1261/rna.080140.124. RNA. 2024. PMID: 39060168
-
Single-molecule visualization of mRNA circularization during translation.Exp Mol Med. 2023 Feb;55(2):283-289. doi: 10.1038/s12276-023-00933-1. Epub 2023 Jan 31. Exp Mol Med. 2023. PMID: 36720916 Free PMC article. Review.
-
Spying on SARS-CoV-2 with Fluorescent Tags and Protease Reporters.Viruses. 2023 Sep 27;15(10):2005. doi: 10.3390/v15102005. Viruses. 2023. PMID: 37896782 Free PMC article.
-
Pharmacological modulators of epithelial immunity uncovered by synthetic genetic tracing of SARS-CoV-2 infection responses.Sci Adv. 2023 Jun 23;9(25):eadf4975. doi: 10.1126/sciadv.adf4975. Epub 2023 Jun 21. Sci Adv. 2023. PMID: 37343108 Free PMC article.
-
Involvement of ribosomal protein L17 and Y-box binding protein 1 in the assembly of hepatitis C virus potentially via their interaction with the 3' untranslated region of the viral genome.J Virol. 2024 Jul 23;98(7):e0052224. doi: 10.1128/jvi.00522-24. Epub 2024 Jun 20. J Virol. 2024. PMID: 38899899 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

