UV-B irradiation-activated E3 ligase GmILPA1 modulates gibberellin catabolism to increase plant height in soybean
- PMID: 37805547
- PMCID: PMC10560287
- DOI: 10.1038/s41467-023-41824-3
UV-B irradiation-activated E3 ligase GmILPA1 modulates gibberellin catabolism to increase plant height in soybean
Abstract
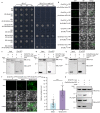
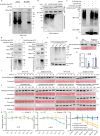
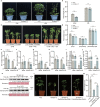
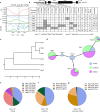
Plant height is a key agronomic trait that affects yield and is controlled by both phytohormone gibberellin (GA) and ultraviolet-B (UV-B) irradiation. However, whether and how plant height is modulated by UV-B-mediated changes in GA metabolism are not well understood. It has not been reported that the E3 ubiquitin ligase Anaphase Promoting Complex/Cyclosome (APC/C) is involved in the regulation of plant growth in response to environmental factors. We perform a forward genetic screen in soybean and find that a mutation in Glycine max Increased Leaf Petiole Angle1 (GmILPA1), encoding a subunit of the APC/C, lead to dwarfism under UV-B irradiation. UV-B promotes the accumulation of GmILPA1, which ubiquitinate the GA catabolic enzyme GA2 OXIDASE-like (GmGA2ox-like), resulting in its degradation in a UV-B-dependent manner. Another E3 ligase, GmUBL1, also ubiquitinate GmGA2ox-like and enhance the GmILPA1-mediated degradation of GmGA2ox-like, which suggest that GmILPA1-GmGA2ox-like module counteract the UV-B-mediated reduction of bioactive GAs. We also determine that GmILPA1 is a target of selection during soybean domestication and breeding. The deletion (Indel-665) in the promoter might facilitate the adaptation of soybean to high UV-B irradiation. This study indicates that an evolutionary GmILPA1 variant has the capability to develop ideal plant architecture with soybean cultivars.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
GmILPA1, Encoding an APC8-like Protein, Controls Leaf Petiole Angle in Soybean.Plant Physiol. 2017 Jun;174(2):1167-1176. doi: 10.1104/pp.16.00074. Epub 2017 Mar 23. Plant Physiol. 2017. PMID: 28336772 Free PMC article.
-
Dysfunction of an Anaphase-Promoting Complex Subunit 8 Homolog Leads to Super-Short Petioles and Enlarged Petiole Angles in Soybean.Int J Mol Sci. 2023 Jul 3;24(13):11024. doi: 10.3390/ijms241311024. Int J Mol Sci. 2023. PMID: 37446203 Free PMC article.
-
GhAPC8 regulates leaf blade angle by modulating multiple hormones in cotton (Gossypium hirsutum L.).Int J Biol Macromol. 2022 Jan 15;195:217-228. doi: 10.1016/j.ijbiomac.2021.11.205. Epub 2021 Dec 9. Int J Biol Macromol. 2022. PMID: 34896470
-
Gibberellin in plant height control: old player, new story.Plant Cell Rep. 2017 Mar;36(3):391-398. doi: 10.1007/s00299-017-2104-5. Epub 2017 Feb 3. Plant Cell Rep. 2017. PMID: 28160061 Review.
-
Functional characterization of Anaphase Promoting Complex/Cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis.Biochim Biophys Acta. 2014 Apr;1845(2):277-93. doi: 10.1016/j.bbcan.2014.02.001. Epub 2014 Feb 22. Biochim Biophys Acta. 2014. PMID: 24569229 Free PMC article. Review.
Cited by
-
Integrating dynamic high-throughput phenotyping and genetic analysis to monitor growth variation in foxtail millet.Plant Methods. 2024 Nov 5;20(1):168. doi: 10.1186/s13007-024-01295-z. Plant Methods. 2024. PMID: 39497091 Free PMC article.
-
Soybean reduced internode 1 determines internode length and improves grain yield at dense planting.Nat Commun. 2023 Dec 1;14(1):7939. doi: 10.1038/s41467-023-42991-z. Nat Commun. 2023. PMID: 38040709 Free PMC article.
-
QTL mapping for plant height and ear height using bi-parental immortalized heterozygous populations in maize.Front Plant Sci. 2024 Mar 25;15:1371394. doi: 10.3389/fpls.2024.1371394. eCollection 2024. Front Plant Sci. 2024. PMID: 38590752 Free PMC article.
References
-
- Peng J, et al. Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. - PubMed
-
- Khush GS. Green revolution: the way forward. Nat. Rev. Genet. 2001;2:815–822. - PubMed
-
- Wang Z, Tian Z. Genomics progress will facilitate molecular breeding in soybean. Sci. China Life Sci. 2015;588:13–815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

