Mesenchymal Stromal Cells Facilitate Neutrophil-Trained Immunity by Reprogramming Hematopoietic Stem Cells
- PMID: 37797588
- PMCID: PMC10622164
- DOI: 10.1159/000533732
Mesenchymal Stromal Cells Facilitate Neutrophil-Trained Immunity by Reprogramming Hematopoietic Stem Cells
Abstract
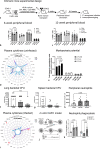
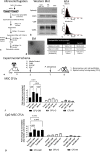
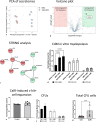
Novel therapeutics are urgently needed to prevent opportunistic infections in immunocompromised individuals undergoing cancer treatments or other immune-suppressive therapies. Trained immunity is a promising strategy to reduce this burden of disease. We previously demonstrated that mesenchymal stromal cells (MSCs) preconditioned with a class A CpG oligodeoxynucleotide (CpG-ODN), a Toll-like receptor 9 (TLR9) agonist, can augment emergency granulopoiesis in a murine model of neutropenic sepsis. Here, we used a chimeric mouse model to demonstrate that MSCs secrete paracrine factors that act on lineage-negative c-kit+ hematopoietic stem cells (HSCs), leaving them "poised" to enhance emergency granulopoiesis months after transplantation. Chimeric mice developed from HSCs exposed to conditioned media from MSCs and CpG-ODN-preconditioned MSCs showed significantly higher bacterial clearance and increased neutrophil granulopoiesis following lung infection than control mice. By Cleavage Under Targets and Release Using Nuclease (CUT&RUN) chromatin sequencing, we identified that MSC-conditioned media leaves H3K4me3 histone marks in HSCs at genes involved in myelopoiesis and in signaling persistence by the mTOR pathway. Both soluble factors and extracellular vesicles from MSCs mediated these effects on HSCs and proteomic analysis by mass spectrometry revealed soluble calreticulin as a potential mediator. In summary, this study demonstrates that trained immunity can be mediated by paracrine factors from MSCs to induce neutrophil-trained immunity by reprogramming HSCs for long-lasting functional changes in neutrophil-mediated antimicrobial immunity.
Keywords: Epigenetics; Hematopoietic stem cells; Neutrophils; Toll-like receptor 9; Trained immunity.
© 2023 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Augmenting emergency granulopoiesis with CpG conditioned mesenchymal stromal cells in murine neutropenic sepsis.Blood Adv. 2020 Oct 13;4(19):4965-4979. doi: 10.1182/bloodadvances.2020002556. Blood Adv. 2020. PMID: 33049055 Free PMC article.
-
Mesenchymal stromal cells enhance the engraftment of hematopoietic stem cells in an autologous mouse transplantation model.Stem Cell Res Ther. 2015 Sep 7;6(1):165. doi: 10.1186/s13287-015-0155-5. Stem Cell Res Ther. 2015. PMID: 26345192 Free PMC article.
-
Mesenchymal stromal cell-derived extracellular vesicles as cell-free biologics for the ex vivo expansion of hematopoietic stem cells.Cell Biol Int. 2020 May;44(5):1078-1102. doi: 10.1002/cbin.11313. Epub 2020 Feb 13. Cell Biol Int. 2020. PMID: 32009258 Review.
-
In vivo monitoring of dynamic interaction between neutrophil and human umbilical cord blood-derived mesenchymal stem cell in mouse liver during sepsis.Stem Cell Res Ther. 2020 Feb 3;11(1):44. doi: 10.1186/s13287-020-1559-4. Stem Cell Res Ther. 2020. PMID: 32014040 Free PMC article.
-
Mechanistic effects of mesenchymal and hematopoietic stem cells: New therapeutic targets in myocardial infarction.J Cell Biochem. 2018 Jul;119(7):5274-5286. doi: 10.1002/jcb.26637. Epub 2018 Mar 12. J Cell Biochem. 2018. PMID: 29266431 Review.
Cited by
-
The causes and consequences of trained immunity in myeloid cells.Front Immunol. 2024 Apr 11;15:1365127. doi: 10.3389/fimmu.2024.1365127. eCollection 2024. Front Immunol. 2024. PMID: 38665915 Free PMC article. Review.
-
Plasticity and crosstalk of mesenchymal stem cells and macrophages in immunomodulation in sepsis.Front Immunol. 2024 Jan 30;15:1338744. doi: 10.3389/fimmu.2024.1338744. eCollection 2024. Front Immunol. 2024. PMID: 38352879 Free PMC article. Review.
References
-
- GBD 2016 Lower Respiratory Infections Collaborators . Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018 Nov;18(11):1191–210. 10.1016/S1473-3099(18)30310-4. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

