An extra-erythrocyte role of haemoglobin body in chondrocyte hypoxia adaption
- PMID: 37794190
- PMCID: PMC10600011
- DOI: 10.1038/s41586-023-06611-6
An extra-erythrocyte role of haemoglobin body in chondrocyte hypoxia adaption
Erratum in
-
Author Correction: An extra-erythrocyte role of haemoglobin body in chondrocyte hypoxia adaption.Nature. 2024 Oct;634(8034):E7. doi: 10.1038/s41586-024-08057-w. Nature. 2024. PMID: 39322681 Free PMC article. No abstract available.
Abstract
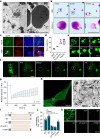
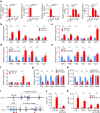
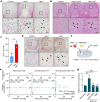
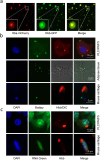
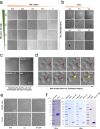
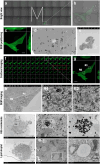
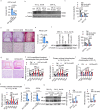
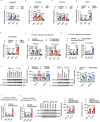
Although haemoglobin is a known carrier of oxygen in erythrocytes that functions to transport oxygen over a long range, its physiological roles outside erythrocytes are largely elusive1,2. Here we found that chondrocytes produced massive amounts of haemoglobin to form eosin-positive bodies in their cytoplasm. The haemoglobin body (Hedy) is a membraneless condensate characterized by phase separation. Production of haemoglobin in chondrocytes is controlled by hypoxia and is dependent on KLF1 rather than the HIF1/2α pathway. Deletion of haemoglobin in chondrocytes leads to Hedy loss along with severe hypoxia, enhanced glycolysis and extensive cell death in the centre of cartilaginous tissue, which is attributed to the loss of the Hedy-controlled oxygen supply under hypoxic conditions. These results demonstrate an extra-erythrocyte role of haemoglobin in chondrocytes, and uncover a heretofore unrecognized mechanism in which chondrocytes survive a hypoxic environment through Hedy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Hypoxia-inducible factor 2alpha is essential for hypoxic induction of the human articular chondrocyte phenotype.Arthritis Rheum. 2007 Oct;56(10):3297-306. doi: 10.1002/art.22878. Arthritis Rheum. 2007. PMID: 17907154
-
Parathyroid hormone-related protein is induced by hypoxia and promotes expression of the differentiated phenotype of human articular chondrocytes.Clin Sci (Lond). 2013 Nov;125(10):461-70. doi: 10.1042/CS20120610. Clin Sci (Lond). 2013. PMID: 23662774
-
Hypoxia. HIF-mediated articular chondrocyte function: prospects for cartilage repair.Arthritis Res Ther. 2009;11(1):213. doi: 10.1186/ar2574. Epub 2009 Feb 5. Arthritis Res Ther. 2009. PMID: 19232075 Free PMC article. Review.
-
Lack of oxygen in articular cartilage: consequences for chondrocyte biology.Int J Exp Pathol. 2010 Apr;91(2):99-106. doi: 10.1111/j.1365-2613.2010.00707.x. Int J Exp Pathol. 2010. PMID: 20384821 Free PMC article. Review.
-
Effects of oxygen on zonal marker expression in human articular chondrocytes.Tissue Eng Part A. 2012 May;18(9-10):920-33. doi: 10.1089/ten.TEA.2011.0088. Epub 2012 Jan 4. Tissue Eng Part A. 2012. PMID: 22097912
Cited by
-
How oxygenation shapes immune responses: emerging roles for physioxia and pathological hypoxia.Nat Rev Immunol. 2024 Sep 30. doi: 10.1038/s41577-024-01087-5. Online ahead of print. Nat Rev Immunol. 2024. PMID: 39349943 Review.
-
Hemoglobin alpha is a redox-sensitive mitochondrial-related protein in T-lymphocytes.bioRxiv [Preprint]. 2024 Sep 20:2024.09.16.613298. doi: 10.1101/2024.09.16.613298. bioRxiv. 2024. PMID: 39345360 Free PMC article. Preprint.
-
A naturally occurring 22-amino acid fragment of human hemoglobin A inhibits autophagy and HIV-1.Cell Mol Life Sci. 2024 Sep 17;81(1):409. doi: 10.1007/s00018-024-05447-1. Cell Mol Life Sci. 2024. PMID: 39289189 Free PMC article.
-
Targeting Parkin-regulated metabolomic change in cartilage in the treatment of osteoarthritis.iScience. 2024 Jul 27;27(9):110597. doi: 10.1016/j.isci.2024.110597. eCollection 2024 Sep 20. iScience. 2024. PMID: 39220257 Free PMC article.
-
Erythroid Krüppel-Like Factor (KLF1): A Surprisingly Versatile Regulator of Erythroid Differentiation.Adv Exp Med Biol. 2024;1459:217-242. doi: 10.1007/978-3-031-62731-6_10. Adv Exp Med Biol. 2024. PMID: 39017846 Review.
References
-
- Semenza, G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer3, 721–732 (2003). - PubMed
-
- Losman, J.-A., Koivunen, P. & Kaelin, W. G. 2-Oxoglutarate-dependent dioxygenases in cancer. Nat. Rev. Cancer20, 710–726 (2020). - PubMed
-
- Gandara, L. & Wappner, P. Metabo-Devo: a metabolic perspective of development. Mech. Dev.154, 12–23 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

