Simultaneous subset tracing and miRNA profiling of tumor-derived exosomes via dual-surface-protein orthogonal barcoding
- PMID: 37792944
- PMCID: PMC10550235
- DOI: 10.1126/sciadv.adi1556
Simultaneous subset tracing and miRNA profiling of tumor-derived exosomes via dual-surface-protein orthogonal barcoding
Abstract
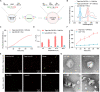
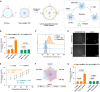
The clinical potential of miRNA-based liquid biopsy has been largely limited by the heterogeneous sources in plasma and tedious assay processes. Here, we develop a precise and robust one-pot assay called dual-surface-protein-guided orthogonal recognition of tumor-derived exosomes and in situ profiling of microRNAs (SORTER) to detect tumor-derived exosomal miRNAs and enhance the diagnostic accuracy of prostate cancer (PCa). The SORTER uses two allosteric aptamers against exosomal marker CD63 and tumor marker EpCAM to create an orthogonal labeling barcode and achieve selective sorting of tumor-specific exosome subtypes. Furthermore, the labeled barcode on tumor-derived exosomes initiated targeted membrane fusion with liposome probes to import miRNA detection reagents, enabling in situ sensitive profiling of tumor-derived exosomal miRNAs. With a signature of six miRNAs, SORTER differentiated PCa and benign prostatic hyperplasia with an accuracy of 100%. Notably, the diagnostic accuracy reached 90.6% in the classification of metastatic and nonmetastatic PCa. We envision that the SORTER will promote the clinical adaptability of miRNA-based liquid biopsy.
Figures





Similar articles
-
Simultaneous multiplexed detection of exosomal microRNAs and surface proteins for prostate cancer diagnosis.Biosens Bioelectron. 2019 Dec 15;146:111749. doi: 10.1016/j.bios.2019.111749. Epub 2019 Oct 1. Biosens Bioelectron. 2019. PMID: 31600625
-
Tumor-Derived Exosomal Long Noncoding RNAs as Promising Diagnostic Biomarkers for Prostate Cancer.Cell Physiol Biochem. 2018;46(2):532-545. doi: 10.1159/000488620. Epub 2018 Mar 26. Cell Physiol Biochem. 2018. PMID: 29614511
-
Exosomal microRNAs in liquid biopsies: future biomarkers for prostate cancer.Clin Transl Oncol. 2017 Jun;19(6):651-657. doi: 10.1007/s12094-016-1599-5. Epub 2017 Jan 4. Clin Transl Oncol. 2017. PMID: 28054319 Review.
-
Hydrogel-based hybridization chain reaction (HCR) for detection of urinary exosomal miRNAs as a diagnostic tool of prostate cancer.Biosens Bioelectron. 2021 Nov 15;192:113504. doi: 10.1016/j.bios.2021.113504. Epub 2021 Jul 16. Biosens Bioelectron. 2021. PMID: 34298498
-
Exosomal noncoding RNAs in prostate cancer.Clin Chim Acta. 2022 Dec 1;537:127-132. doi: 10.1016/j.cca.2022.10.018. Epub 2022 Oct 29. Clin Chim Acta. 2022. PMID: 36330945 Review.
Cited by
-
The new advance of exosome-based liquid biopsy for cancer diagnosis.J Nanobiotechnology. 2024 Oct 8;22(1):610. doi: 10.1186/s12951-024-02863-0. J Nanobiotechnology. 2024. PMID: 39380060 Free PMC article. Review.
-
Hypoxia-Driven Changes in Tumor Microenvironment: Insights into Exosome-Mediated Cell Interactions.Int J Nanomedicine. 2024 Aug 12;19:8211-8236. doi: 10.2147/IJN.S479533. eCollection 2024. Int J Nanomedicine. 2024. PMID: 39157736 Free PMC article. Review.
References
-
- N. Mahtal, O. Lenoir, C. Tinel, D. Anglicheau, P. L. Tharaux, MicroRNAs in kidney injury and disease. Nat. Rev. Nephrol. 18, 643–662 (2022). - PubMed
-
- D. Santovito, C. Weber, Non-canonical features of microRNAs: Paradigms emerging from cardiovascular disease. Nat. Rev. Cardiol. 19, 620–638 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

