Compartmentalization of androgen receptors at endogenous genes in living cells
- PMID: 37791849
- PMCID: PMC10639085
- DOI: 10.1093/nar/gkad803
Compartmentalization of androgen receptors at endogenous genes in living cells
Abstract
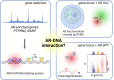
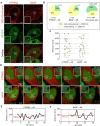
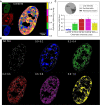
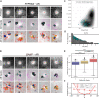
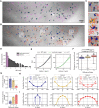
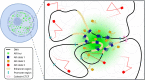
A wide range of nuclear proteins are involved in the spatio-temporal organization of the genome through diverse biological processes such as gene transcription and DNA replication. Upon stimulation by testosterone and translocation to the nucleus, multiple androgen receptors (ARs) accumulate in microscopically discernable foci which are irregularly distributed in the nucleus. Here, we investigated the formation and physical nature of these foci, by combining novel fluorescent labeling techniques to visualize a defined chromatin locus of AR-regulated genes-PTPRN2 or BANP-simultaneously with either AR foci or individual AR molecules. Quantitative colocalization analysis showed evidence of AR foci formation induced by R1881 at both PTPRN2 and BANP loci. Furthermore, single-particle tracking (SPT) revealed three distinct subdiffusive fractional Brownian motion (fBm) states: immobilized ARs were observed near the labeled genes likely as a consequence of DNA-binding, while the intermediate confined state showed a similar spatial behavior but with larger displacements, suggesting compartmentalization by liquid-liquid phase separation (LLPS), while freely mobile ARs were diffusing in the nuclear environment. All together, we show for the first time in living cells the presence of AR-regulated genes in AR foci.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Antiandrogens prevent stable DNA-binding of the androgen receptor.J Cell Sci. 2005 Sep 15;118(Pt 18):4187-98. doi: 10.1242/jcs.02546. Epub 2005 Sep 1. J Cell Sci. 2005. PMID: 16141232
-
Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility.J Cell Biochem. 2006 Jul 1;98(4):770-88. doi: 10.1002/jcb.20593. J Cell Biochem. 2006. PMID: 16440331
-
Role of the promyelocytic leukemia body in the dynamic interaction between the androgen receptor and steroid receptor coactivator-1 in living cells.Mol Endocrinol. 2003 Jan;17(1):128-40. doi: 10.1210/me.2002-0165. Mol Endocrinol. 2003. PMID: 12511612
-
Intranuclear organization and function of the androgen receptor.Trends Endocrinol Metab. 2004 Nov;15(9):411-7. doi: 10.1016/j.tem.2004.09.006. Trends Endocrinol Metab. 2004. PMID: 15519887 Review.
-
Impacts of chromatin dynamics and compartmentalization on DNA repair.DNA Repair (Amst). 2021 Sep;105:103162. doi: 10.1016/j.dnarep.2021.103162. Epub 2021 Jun 19. DNA Repair (Amst). 2021. PMID: 34182258 Review.
Cited by
-
Phase Separation Mediated Sub-Nuclear Compartmentalization of Androgen Receptors.Cells. 2024 Oct 13;13(20):1693. doi: 10.3390/cells13201693. Cells. 2024. PMID: 39451211 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

