Estimating the contribution of CD4 T cell subset proliferation and differentiation to HIV persistence
- PMID: 37783718
- PMCID: PMC10545742
- DOI: 10.1038/s41467-023-41521-1
Estimating the contribution of CD4 T cell subset proliferation and differentiation to HIV persistence
Abstract
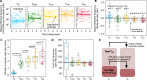
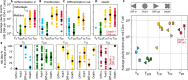
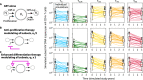
Persistence of HIV in people living with HIV (PWH) on suppressive antiretroviral therapy (ART) has been linked to physiological mechanisms of CD4+ T cells. Here, in the same 37 male PWH on ART we measure longitudinal kinetics of HIV DNA and cell turnover rates in five CD4 cell subsets: naïve (TN), stem-cell- (TSCM), central- (TCM), transitional- (TTM), and effector-memory (TEM). HIV decreases in TTM and TEM but not in less-differentiated subsets. Cell turnover is ~10 times faster than HIV clearance in memory subsets, implying that cellular proliferation consistently creates HIV DNA. The optimal mathematical model for these integrated data sets posits HIV DNA also passages between CD4 cell subsets via cellular differentiation. Estimates are heterogeneous, but in an average participant's year ~10 (in TN and TSCM) and ~104 (in TCM, TTM, TEM) proviruses are generated by proliferation while ~103 proviruses passage via cell differentiation (per million CD4). In simulations, therapies blocking proliferation and/or enhancing differentiation could reduce HIV DNA by 1-2 logs over 3 years. In summary, HIV exploits cellular proliferation and differentiation to persist during ART but clears faster in more proliferative/differentiated CD4 cell subsets and the same physiological mechanisms sustaining HIV might be temporarily modified to reduce it.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Initiation of Antiretroviral Therapy Restores CD4+ T Memory Stem Cell Homeostasis in Simian Immunodeficiency Virus-Infected Macaques.J Virol. 2016 Jul 11;90(15):6699-6708. doi: 10.1128/JVI.00492-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27170752 Free PMC article.
-
Atlas of the HIV-1 Reservoir in Peripheral CD4 T Cells of Individuals on Successful Antiretroviral Therapy.mBio. 2021 Dec 21;12(6):e0307821. doi: 10.1128/mBio.03078-21. Epub 2021 Nov 30. mBio. 2021. PMID: 34844430 Free PMC article.
-
Differentiation into an Effector Memory Phenotype Potentiates HIV-1 Latency Reversal in CD4+ T Cells.J Virol. 2019 Nov 26;93(24):e00969-19. doi: 10.1128/JVI.00969-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31578289 Free PMC article.
-
T memory stem cells and HIV: a long-term relationship.Curr HIV/AIDS Rep. 2015 Mar;12(1):33-40. doi: 10.1007/s11904-014-0246-4. Curr HIV/AIDS Rep. 2015. PMID: 25578055 Free PMC article. Review.
-
Episomal HIV-1 DNA and its relationship to other markers of HIV-1 persistence.Retrovirology. 2018 Jan 30;15(1):15. doi: 10.1186/s12977-018-0398-1. Retrovirology. 2018. PMID: 29378611 Free PMC article. Review.
Cited by
-
Modelling HIV-1 control and remission.NPJ Syst Biol Appl. 2024 Aug 8;10(1):84. doi: 10.1038/s41540-024-00407-8. NPJ Syst Biol Appl. 2024. PMID: 39117718 Free PMC article. Review.
-
Highlights from the Inaugural HIV Reservoirs and Immune Control Conference, October 1st-4th 2023, Malahide Ireland.Pathog Immun. 2023 Dec 19;8(1):161-169. doi: 10.20411/pai.v8i1.653. eCollection 2023. Pathog Immun. 2023. PMID: 38155941 Free PMC article.
-
Interactions Between HDL and CD4+ T Cells: A Novel Understanding of HDL Anti-Inflammatory Properties.Arterioscler Thromb Vasc Biol. 2024 Jun;44(6):1191-1201. doi: 10.1161/ATVBAHA.124.320851. Epub 2024 Apr 25. Arterioscler Thromb Vasc Biol. 2024. PMID: 38660807 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

