Heat-inactivated Bifidobacterium adolescentis ameliorates colon senescence through Paneth-like-cell-mediated stem cell activation
- PMID: 37777508
- PMCID: PMC10542354
- DOI: 10.1038/s41467-023-41827-0
Heat-inactivated Bifidobacterium adolescentis ameliorates colon senescence through Paneth-like-cell-mediated stem cell activation
Abstract
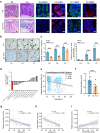
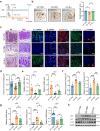
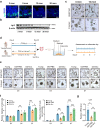
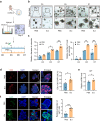
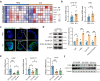
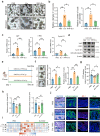
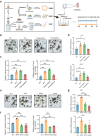
Declined numbers and weakened functions of intestinal stem cells (ISCs) impair the integrity of the intestinal epithelium during aging. However, the impact of intestinal microbiota on ISCs in this process is unclear. Here, using premature aging mice (telomerase RNA component knockout, Terc-/-), natural aging mice, and in vitro colonoid models, we explore how heat-inactivated Bifidobacterium adolescentis (B. adolescentis) affects colon senescence. We find that B. adolescentis could mitigate colonic senescence-related changes by enhancing intestinal integrity and stimulating the regeneration of Lgr5+ ISCs via Wnt/β-catenin signaling. Furthermore, we uncover the involvement of Paneth-like cells (PLCs) within the colonic stem-cell-supporting niche in the B. adolescentis-induced ISC regeneration. In addition, we identify soluble polysaccharides (SPS) as potential effective components of B. adolescentis. Overall, our findings reveal the role of heat-inactivated B. adolescentis in maintaining the ISCs regeneration and intestinal barrier, and propose a microbiota target for ameliorating colon senescence.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Bifidobacterium adolescentis Isolated from Different Hosts Modifies the Intestinal Microbiota and Displays Differential Metabolic and Immunomodulatory Properties in Mice Fed a High-Fat Diet.Nutrients. 2021 Mar 21;13(3):1017. doi: 10.3390/nu13031017. Nutrients. 2021. PMID: 33801119 Free PMC article.
-
Canonical Wnt Signaling Ameliorates Aging of Intestinal Stem Cells.Cell Rep. 2017 Mar 14;18(11):2608-2621. doi: 10.1016/j.celrep.2017.02.056. Cell Rep. 2017. PMID: 28297666 Free PMC article.
-
Simultaneous real-time analysis of Paneth cell and intestinal stem cell response to interferon-γ by a novel stem cell niche tracking method.Biochem Biophys Res Commun. 2021 Mar 19;545:14-19. doi: 10.1016/j.bbrc.2021.01.050. Epub 2021 Jan 30. Biochem Biophys Res Commun. 2021. PMID: 33529805
-
Plasticity of Intestinal Epithelium: Stem Cell Niches and Regulatory Signals.Int J Mol Sci. 2020 Dec 31;22(1):357. doi: 10.3390/ijms22010357. Int J Mol Sci. 2020. PMID: 33396437 Free PMC article. Review.
-
Molecular regulation mechanism of intestinal stem cells in mucosal injury and repair in ulcerative colitis.World J Gastroenterol. 2023 Apr 28;29(16):2380-2396. doi: 10.3748/wjg.v29.i16.2380. World J Gastroenterol. 2023. PMID: 37179583 Free PMC article. Review.
Cited by
-
Metastasis of colon cancer requires Dickkopf-2 to generate cancer cells with Paneth cell properties.bioRxiv [Preprint]. 2024 Sep 25:2024.04.12.589235. doi: 10.1101/2024.04.12.589235. bioRxiv. 2024. Update in: Elife. 2024 Nov 13;13:RP97279. doi: 10.7554/eLife.97279 PMID: 38659853 Free PMC article. Updated. Preprint.
-
The connection between aging, cellular senescence and gut microbiome alterations: A comprehensive review.Aging Cell. 2024 Oct;23(10):e14315. doi: 10.1111/acel.14315. Epub 2024 Aug 15. Aging Cell. 2024. PMID: 39148278 Free PMC article. Review.
-
Metastasis of colon cancer requires Dickkopf-2 to generate cancer cells with Paneth cell properties.Elife. 2024 Nov 13;13:RP97279. doi: 10.7554/eLife.97279. Elife. 2024. PMID: 39535280 Free PMC article.
-
L-arginine metabolism ameliorates age-related cognitive impairment by Amuc_1100-mediated gut homeostasis maintaining.Aging Cell. 2024 Apr;23(4):e14081. doi: 10.1111/acel.14081. Epub 2024 Jan 18. Aging Cell. 2024. PMID: 38236004 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

