E3 ubiquitin ligase TRIM21 targets TIF1γ to regulate β-catenin signaling in glioblastoma
- PMID: 37771771
- PMCID: PMC10526654
- DOI: 10.7150/thno.85662
E3 ubiquitin ligase TRIM21 targets TIF1γ to regulate β-catenin signaling in glioblastoma
Abstract
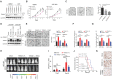
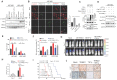
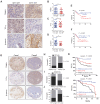
Background: Elucidation of the mechanism of ubiquitation has led to novel ways to treat glioblastoma (GBM). A tripartite motif (TRIM) protein mediates a reversible, stringent ubiquitation which is closely related to glioma malignancy. This study intends to screen the most vital and abnormal regulating component of the tripartite motif protein and to explore its underlying mechanisms. Methods: TRIM21 is identified as an important oncogene that accelerates the progression of glioma cell through database in a multidimensional way and this is confirmed in human samples and cells. Tandem Mass Tags (TMT) and MS analysis are performed to discover the substrates of TRIM21.The underlying mechanisms are further investigated by CO-IP, luciferase reporter assays and gain and loss of function assays. In vivo treatment with siRNA is applied to evaluate the therapeutic significance of TRIM21. Result: We screened a panel of TRIM proteins and identified TRIM21, a E3 ubiquitin-protein ligase and autoantigen, as well as a prognostic biomarker for GBM. Functionally, high expression of wild-type TRIM21 accelerates tumor progression in vitro and in vivo, whereas TRIM21 mutants, including one with a critical RING-finger deletion, do not. Mechanistically, TRIM21 stimulates K63-linked ubiquitination and subcellular translocation of active β-catenin from the cytoplasm to the nucleus. Moreover, TRIM21 forms a complex with the β-catenin upstream regulator, TIF1γ, in the nucleus and accelerated its degradation by inducing K48-linked ubiquitination at K5 site, consequently increasing further nuclear β-catenin presence. Endogenous TRIM21 levels are found to be inversely correlated with TIF1γ but positively correlated with β-catenin in glioma tissue microarray experiments. Furthermore, direct injection of TRIM21 small interfering RNA (siRNA) into U87 cell-derived tumors (in vivo treatment with siRNA) is proved to inhibit tumor growth in nude mice. Conclusion: This work suggests that TRIM21/TIF1γ/β-catenin axis is involved in the progression of human GBM. TRIM21 is a promising therapeutic and prognostic biomarker for glioma with hyperactive β-catenin.
Keywords: TIF1γ; TRIM21; TRIMs family; ubiquitination; β-catenin.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




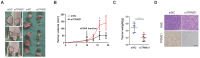
Similar articles
-
TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα.Cell Death Differ. 2021 Jan;28(1):367-381. doi: 10.1038/s41418-020-00606-w. Epub 2020 Aug 19. Cell Death Differ. 2021. PMID: 32814880 Free PMC article.
-
The E3 ubiquitin ligase HUWE1 acts through the N-Myc-DLL1-NOTCH1 signaling axis to suppress glioblastoma progression.Cancer Commun (Lond). 2022 Sep;42(9):868-886. doi: 10.1002/cac2.12334. Epub 2022 Jul 18. Cancer Commun (Lond). 2022. PMID: 35848447 Free PMC article.
-
The deubiquitinase USP7 and E3 ligase TRIM21 regulate vasculogenic mimicry and malignant progression of RMS by balancing SNAI2 homeostasis.J Exp Clin Cancer Res. 2024 May 4;43(1):135. doi: 10.1186/s13046-024-03056-1. J Exp Clin Cancer Res. 2024. PMID: 38702792 Free PMC article.
-
Multiple Roles of TRIM21 in Virus Infection.Int J Mol Sci. 2023 Jan 14;24(2):1683. doi: 10.3390/ijms24021683. Int J Mol Sci. 2023. PMID: 36675197 Free PMC article. Review.
-
The molecular mechanisms that drive intracellular neutralization by the antibody-receptor and RING E3 ligase TRIM21.Semin Cell Dev Biol. 2022 Jun;126:99-107. doi: 10.1016/j.semcdb.2021.11.005. Epub 2021 Nov 22. Semin Cell Dev Biol. 2022. PMID: 34823983 Review.
Cited by
-
Neddylation activated TRIM25 desensitizes triple-negative breast cancer to paclitaxel via TFEB-mediated autophagy.J Exp Clin Cancer Res. 2024 Jun 26;43(1):177. doi: 10.1186/s13046-024-03085-w. J Exp Clin Cancer Res. 2024. PMID: 38926803 Free PMC article.
-
Regulation of CD73 on NAD metabolism: Unravelling the interplay between tumour immunity and tumour metabolism.Cell Commun Signal. 2024 Aug 1;22(1):387. doi: 10.1186/s12964-024-01755-y. Cell Commun Signal. 2024. PMID: 39090604 Free PMC article. Review.
-
Multifaceted role of TRIM28 in health and disease.MedComm (2020). 2024 Nov 11;5(11):e790. doi: 10.1002/mco2.790. eCollection 2024 Nov. MedComm (2020). 2024. PMID: 39534556 Free PMC article. Review.
-
Emerging roles of tripartite motif family proteins (TRIMs) in breast cancer.Cancer Med. 2024 Jul;13(14):e7472. doi: 10.1002/cam4.7472. Cancer Med. 2024. PMID: 39016065 Free PMC article. Review.
-
Erianin induces ferroptosis in GSCs via REST/LRSAM1 mediated SLC40A1 ubiquitination to overcome TMZ resistance.Cell Death Dis. 2024 Jul 22;15(7):522. doi: 10.1038/s41419-024-06902-4. Cell Death Dis. 2024. PMID: 39039049 Free PMC article.
References
-
- Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. - PubMed
-
- Jin WL, Mao XY, Qiu GZ. Targeting Deubiquitinating Enzymes in Glioblastoma Multiforme: Expectations and Challenges. Med Res Rev. 2017;37:627–661. - PubMed
-
- Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol. 2012;770:27–37. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

