Crystal structures and molecular dynamics simulations of a humanised antibody fragment at acidic to basic pH
- PMID: 37770469
- PMCID: PMC10539359
- DOI: 10.1038/s41598-023-42698-7
Crystal structures and molecular dynamics simulations of a humanised antibody fragment at acidic to basic pH
Abstract
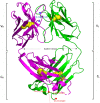
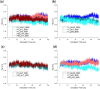
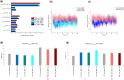
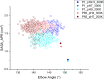
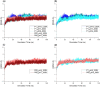
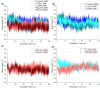
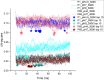
Antibody-fragment (Fab) therapy development has the potential to be accelerated by computational modelling and simulations that predict their target binding, stability, formulation, manufacturability, and the impact of further protein engineering. Such approaches are currently predicated on starting with good crystal structures that closely represent those found under the solution conditions to be simulated. A33 Fab, is an undeveloped immunotherapeutic antibody candidate that was targeted to the human A33 antigen homogeneously expressed in 95% cases of primary and metastatic colorectal cancers. It is now used as a very well characterised testing ground for developing analytics, formulation and protein engineering strategies, and to gain a deeper understanding of mechanisms of destabilisation, representative of the wider therapeutic Fab platform. In this article, we report the structure of A33 Fab in two different crystal forms obtained at acidic and basic pH. The structures overlapped with RMSD of 1.33 Å overall, yet only 0.5 Å and 0.76 Å for the variable- and constant regions alone. While most of the differences were within experimental error, the switch linker between the variable and the constant regions showed some small differences between the two pHs. The two structures then enabled a direct evaluation of the impact of initial crystal structure selection on the outcomes of molecular dynamics simulations under different conditions, and their subsequent use for determining best fit solution structures using previously obtained small-angle x-ray scattering (SAXS) data. The differences in the two structures did not have a major impact on MD simulations regardless of the pH, other than a slight persistence of structure affecting the solvent accessibility of one of the predicted APR regions of A33 Fab. Interestingly, despite being obtained at pH 4 and pH 9, the two crystal structures were more similar to the SAXS solution structures obtained at pH 7, than to those at pH 4 or pH 9. Furthermore, the P65 crystal structure from pH 4 was also a better representation of the solution structures at any other pH, than was the P1 structure obtained at pH 9. Thus, while obtained at different pH, the two crystal structures may represent highly (P65) and lesser (P1) populated species that both exist at pH 7 in solution. These results now lay the foundation for confident MD simulations of A33 Fab that rationalise or predict behaviours in a range of conditions.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
An Expanded Conformation of an Antibody Fab Region by X-Ray Scattering, Molecular Dynamics, and smFRET Identifies an Aggregation Mechanism.J Mol Biol. 2019 Mar 29;431(7):1409-1425. doi: 10.1016/j.jmb.2019.02.009. Epub 2019 Feb 16. J Mol Biol. 2019. PMID: 30776431
-
Evaluating Anti-CD32b F(ab) Conformation Using Molecular Dynamics and Small-Angle X-Ray Scattering.Biophys J. 2018 Jul 17;115(2):289-299. doi: 10.1016/j.bpj.2018.03.040. Biophys J. 2018. PMID: 30021105 Free PMC article.
-
Hybrid Methods for Modeling Protein Structures Using Molecular Dynamics Simulations and Small-Angle X-Ray Scattering Data.Adv Exp Med Biol. 2018;1105:237-258. doi: 10.1007/978-981-13-2200-6_15. Adv Exp Med Biol. 2018. PMID: 30617833 Review.
-
Characterization of the NISTmAb Reference Material using small-angle scattering and molecular simulation : Part I: Dilute protein solutions.Anal Bioanal Chem. 2018 Mar;410(8):2141-2159. doi: 10.1007/s00216-018-0868-2. Epub 2018 Feb 9. Anal Bioanal Chem. 2018. PMID: 29423600
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
References
-
- King, D. J., Adair, J. R. & Owens, R. J. (Google Patents, 2001).
-
- Welt S, et al. Phase I study of anticolon cancer humanized antibody A33. Clin. Cancer Res. 2003;9:1338–1346. - PubMed
-
- GarinChesa P, et al. Organ-specific expression of the colon cancer antigen A33, a cell surface target for antibody-based therapy. Int. J. Oncol. 1996;9:465–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

