Context-dependent function of the transcriptional regulator Rap1 in gene silencing and activation in Saccharomyces cerevisiae
- PMID: 37769255
- PMCID: PMC10556627
- DOI: 10.1073/pnas.2304343120
Context-dependent function of the transcriptional regulator Rap1 in gene silencing and activation in Saccharomyces cerevisiae
Abstract
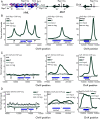
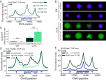
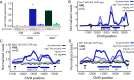
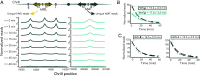
In Saccharomyces cerevisiae, heterochromatin is formed through interactions between site-specific DNA-binding factors, including the transcriptional activator Repressor Activator Protein (Rap1), and Sir proteins. Despite an understanding of the establishment and maintenance of Sir-silenced chromatin, the mechanism of gene silencing by Sir proteins has remained a mystery. Utilizing high-resolution chromatin immunoprecipitation, we found that Rap1, the native activator of the bidirectional HMLα promoter, bound its recognition sequence in silenced chromatin, and its binding was enhanced by the presence of Sir proteins. In contrast to prior results, various components of transcription machinery were not able to access HMLα in the silenced state. These findings disproved the long-standing model of indiscriminate steric occlusion by Sir proteins and led to investigation of the role of the transcriptional activator Rap1 in Sir-silenced chromatin. Using a highly sensitive assay that monitors loss-of-silencing events, we identified a role for promoter-bound Rap1 in the maintenance of silent chromatin through interactions with the Sir complex. We also found that promoter-bound Rap1 activated HMLα when in an expressed state, and aided in the transition from transcription initiation to elongation. Highlighting the importance of epigenetic context in transcription factor function, these results point toward a model in which the duality of Rap1 function was mediated by local chromatin environment rather than binding-site availability.
Keywords: chromatin; epigenetics; gene silencing.
Conflict of interest statement
The authors declare no competing interest.
Figures
Update of
-
Context dependent function of the transcriptional regulator Rap1 in gene silencing and activation in Saccharomyces cerevisiae.bioRxiv [Preprint]. 2023 May 11:2023.05.08.539937. doi: 10.1101/2023.05.08.539937. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Oct 3;120(40):e2304343120. doi: 10.1073/pnas.2304343120. PMID: 37214837 Free PMC article. Updated. Preprint.
Similar articles
-
Context dependent function of the transcriptional regulator Rap1 in gene silencing and activation in Saccharomyces cerevisiae.bioRxiv [Preprint]. 2023 May 11:2023.05.08.539937. doi: 10.1101/2023.05.08.539937. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Oct 3;120(40):e2304343120. doi: 10.1073/pnas.2304343120. PMID: 37214837 Free PMC article. Updated. Preprint.
-
Functions of protosilencers in the formation and maintenance of heterochromatin in Saccharomyces cerevisiae.PLoS One. 2012;7(5):e37092. doi: 10.1371/journal.pone.0037092. Epub 2012 May 17. PLoS One. 2012. PMID: 22615905 Free PMC article.
-
On the Mechanism of Gene Silencing in Saccharomyces cerevisiae.G3 (Bethesda). 2015 Jun 16;5(8):1751-63. doi: 10.1534/g3.115.018515. G3 (Bethesda). 2015. PMID: 26082137 Free PMC article.
-
Transcriptional silencing in Saccharomyces cerevisiae: known unknowns.Epigenetics Chromatin. 2024 Sep 14;17(1):28. doi: 10.1186/s13072-024-00553-7. Epigenetics Chromatin. 2024. PMID: 39272151 Free PMC article. Review.
-
SIR proteins and the assembly of silent chromatin in budding yeast.Annu Rev Genet. 2013;47:275-306. doi: 10.1146/annurev-genet-021313-173730. Epub 2013 Sep 4. Annu Rev Genet. 2013. PMID: 24016189 Review.
Cited by
-
ChEC-seq2: an improved Chromatin Endogenous Cleavage sequencing method and bioinformatic analysis pipeline for mapping in vivo protein-DNA interactions.bioRxiv [Preprint]. 2023 Oct 18:2023.10.15.562421. doi: 10.1101/2023.10.15.562421. bioRxiv. 2023. Update in: NAR Genom Bioinform. 2024 Feb 07;6(1):lqae012. doi: 10.1093/nargab/lqae012. PMID: 37905156 Free PMC article. Updated. Preprint.
-
ChEC-seq2: an improved chromatin endogenous cleavage sequencing method and bioinformatic analysis pipeline for mapping in vivo protein-DNA interactions.NAR Genom Bioinform. 2024 Feb 7;6(1):lqae012. doi: 10.1093/nargab/lqae012. eCollection 2024 Mar. NAR Genom Bioinform. 2024. PMID: 38327869 Free PMC article.
-
Sir2 and Fun30 regulate ribosomal DNA replication timing via MCM helicase positioning and nucleosome occupancy.bioRxiv [Preprint]. 2024 Oct 28:2024.03.21.586113. doi: 10.1101/2024.03.21.586113. bioRxiv. 2024. Update in: Elife. 2025 Jan 20;13:RP97438. doi: 10.7554/eLife.97438. PMID: 38585982 Free PMC article. Updated. Preprint.
References
-
- Rusche L. N., Kirchmaier A. L., Rine J., The establishment, inheritance, and function of silenced chromatin in saccharomyces cerevisiae. Annu. Rev. Biochem. 72, 481–516 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

