Unraveling the Dynamics of Omicron (BA.1, BA.2, and BA.5) Waves and Emergence of the Deltacton Variant: Genomic Epidemiology of the SARS-CoV-2 Epidemic in Cyprus (Oct 2021-Oct 2022)
- PMID: 37766339
- PMCID: PMC10535466
- DOI: 10.3390/v15091933
Unraveling the Dynamics of Omicron (BA.1, BA.2, and BA.5) Waves and Emergence of the Deltacton Variant: Genomic Epidemiology of the SARS-CoV-2 Epidemic in Cyprus (Oct 2021-Oct 2022)
Abstract
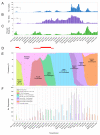
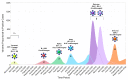
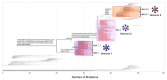
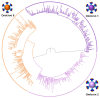
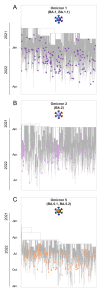
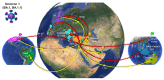
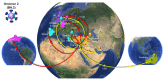
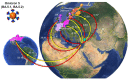
Commencing in December 2019 with the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), three years of the coronavirus disease 2019 (COVID-19) pandemic have transpired. The virus has consistently demonstrated a tendency for evolutionary adaptation, resulting in mutations that impact both immune evasion and transmissibility. This ongoing process has led to successive waves of infections. This study offers a comprehensive assessment spanning genetic, phylogenetic, phylodynamic, and phylogeographic dimensions, focused on the trajectory of the SARS-CoV-2 epidemic in Cyprus. Based on a dataset comprising 4700 viral genomic sequences obtained from affected individuals between October 2021 and October 2022, our analysis is presented. Over this timeframe, a total of 167 distinct lineages and sublineages emerged, including variants such as Delta and Omicron (1, 2, and 5). Notably, during the fifth wave of infections, Omicron subvariants 1 and 2 gained prominence, followed by the ascendancy of Omicron 5 in the subsequent sixth wave. Additionally, during the fifth wave (December 2021-January 2022), a unique set of Delta sequences with genetic mutations associated with Omicron variant 1, dubbed "Deltacron", was identified. The emergence of this phenomenon initially evoked skepticism, characterized by concerns primarily centered around contamination or coinfection as plausible etiological contributors. These hypotheses were predominantly disseminated through unsubstantiated assertions within the realms of social and mass media, lacking concurrent scientific evidence to validate their claims. Nevertheless, the exhaustive molecular analyses presented in this study have demonstrated that such occurrences would likely lead to a frameshift mutation-a genetic aberration conspicuously absent in our provided sequences. This substantiates the accuracy of our initial assertion while refuting contamination or coinfection as potential etiologies. Comparable observations on a global scale dispelled doubt, eventually leading to the recognition of Delta-Omicron variants by the scientific community and their subsequent monitoring by the World Health Organization (WHO). As our investigation delved deeper into the intricate dynamics of the SARS-CoV-2 epidemic in Cyprus, a discernible pattern emerged, highlighting the major role of international connections in shaping the virus's local trajectory. Notably, the United States and the United Kingdom were the central conduits governing the entry and exit of the virus to and from Cyprus. Moreover, notable migratory routes included nations such as Greece, South Korea, France, Germany, Brazil, Spain, Australia, Denmark, Sweden, and Italy. These empirical findings underscore that the spread of SARS-CoV-2 within Cyprus was markedly influenced by the influx of new, highly transmissible variants, triggering successive waves of infection. This investigation elucidates the emergence of new waves of infection subsequent to the advent of highly contagious and transmissible viral variants, notably characterized by an abundance of mutations localized within the spike protein. Notably, this discovery decisively contradicts the hitherto hypothesis of seasonal fluctuations in the virus's epidemiological dynamics. This study emphasizes the importance of meticulously examining molecular genetics alongside virus migration patterns within a specific region. Past experiences also emphasize the substantial evolutionary potential of viruses such as SARS-CoV-2, underscoring the need for sustained vigilance. However, as the pandemic's dynamics continue to evolve, a balanced approach between caution and resilience becomes paramount. This ethos encourages an approach founded on informed prudence and self-preservation, guided by public health authorities, rather than enduring apprehension. Such an approach empowers societies to adapt and progress, fostering a poised confidence rooted in well-founded adaptation.
Keywords: COVID-19; Cyprus; SARS-CoV-2; genomic epidemiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genomic Epidemiology of the SARS-CoV-2 Epidemic in Cyprus from November 2020 to October 2021: The Passage of Waves of Alpha and Delta Variants of Concern.Viruses. 2022 Dec 30;15(1):108. doi: 10.3390/v15010108. Viruses. 2022. PMID: 36680148 Free PMC article.
-
A Comprehensive Molecular Epidemiological Analysis of SARS-CoV-2 Infection in Cyprus from April 2020 to January 2021: Evidence of a Highly Polyphyletic and Evolving Epidemic.Viruses. 2021 Jun 9;13(6):1098. doi: 10.3390/v13061098. Viruses. 2021. PMID: 34207490 Free PMC article.
-
Spatiotemporal dynamics and epidemiological impact of SARS-CoV-2 XBB lineage dissemination in Brazil in 2023.Microbiol Spectr. 2024 Mar 5;12(3):e0383123. doi: 10.1128/spectrum.03831-23. Epub 2024 Feb 5. Microbiol Spectr. 2024. PMID: 38315011 Free PMC article.
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Omicron variant (B.1.1.529) and its sublineages: What do we know so far amid the emergence of recombinant variants of SARS-CoV-2?Biomed Pharmacother. 2022 Oct;154:113522. doi: 10.1016/j.biopha.2022.113522. Epub 2022 Aug 15. Biomed Pharmacother. 2022. PMID: 36030585 Free PMC article. Review.
Cited by
-
Severity of Omicron Subvariants and Vaccine Impact in Catalonia, Spain.Vaccines (Basel). 2024 Apr 27;12(5):466. doi: 10.3390/vaccines12050466. Vaccines (Basel). 2024. PMID: 38793717 Free PMC article.
-
Factors Associated with SARS-CoV-2 Infection in Fully Vaccinated Nursing Home Residents and Workers.Viruses. 2024 Jan 25;16(2):186. doi: 10.3390/v16020186. Viruses. 2024. PMID: 38399962 Free PMC article.
References
-
- Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). COVID-19 Dashboard. [(accessed on 15 August 2023)]. Available online: https://gisanddata.maps.arcgis.com/apps/dashboards/bda7594740fd402994234....
-
- The World Health Organization (WHO) Tracking SARS-CoV-2 Variants. [(accessed on 3 May 2023)]. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants.
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

