Aldehyde Dehydrogenase Genes as Prospective Actionable Targets in Acute Myeloid Leukemia
- PMID: 37761947
- PMCID: PMC10531322
- DOI: 10.3390/genes14091807
Aldehyde Dehydrogenase Genes as Prospective Actionable Targets in Acute Myeloid Leukemia
Abstract
It has been previously shown that the aldehyde dehydrogenase (ALDH) family member ALDH1A1 has a significant association with acute myeloid leukemia (AML) patient risk group classification and that AML cells lacking ALDH1A1 expression can be readily killed via chemotherapy. In the past, however, a redundancy between the activities of subgroup members of the ALDH family has hampered the search for conclusive evidence to address the role of specific ALDH genes. Here, we describe the bioinformatics evaluation of all nineteen member genes of the ALDH family as prospective actionable targets for the development of methods aimed to improve AML treatment. We implicate ALDH1A1 in the development of recurrent AML, and we show that from the nineteen members of the ALDH family, ALDH1A1 and ALDH2 have the strongest association with AML patient risk group classification. Furthermore, we discover that the sum of the expression values for RNA from the genes, ALDH1A1 and ALDH2, has a stronger association with AML patient risk group classification and survival than either one gene alone does. In conclusion, we identify ALDH1A1 and ALDH2 as prospective actionable targets for the treatment of AML in high-risk patients. Substances that inhibit both enzymatic activities constitute potentially effective pharmaceutics.
Keywords: acute; aldehyde dehydrogenase; biomarkers; cancer bioinformatics; gene expression; leukemia; myeloid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
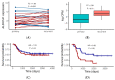
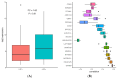
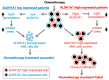
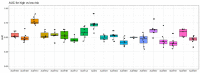
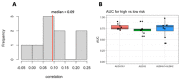

Similar articles
-
Investigating the biology of microRNA links to ALDH1A1 reveals candidates for preclinical testing in acute myeloid leukemia.Int J Oncol. 2024 Dec;65(6):115. doi: 10.3892/ijo.2024.5703. Epub 2024 Nov 8. Int J Oncol. 2024. PMID: 39513593 Free PMC article. Review.
-
Lower RNA expression of ALDH1A1 distinguishes the favorable risk group in acute myeloid leukemia.Mol Biol Rep. 2022 Apr;49(4):3321-3331. doi: 10.1007/s11033-021-07073-7. Epub 2022 Jan 14. Mol Biol Rep. 2022. PMID: 35028852 Review.
-
The Molecular Context of Oxidant Stress Response in Cancer Establishes ALDH1A1 as a Critical Target: What This Means for Acute Myeloid Leukemia.Int J Mol Sci. 2023 May 27;24(11):9372. doi: 10.3390/ijms24119372. Int J Mol Sci. 2023. PMID: 37298333 Free PMC article. Review.
-
Bone marrow stromal cells induce an ALDH+ stem cell-like phenotype and enhance therapy resistance in AML through a TGF-β-p38-ALDH2 pathway.PLoS One. 2020 Nov 30;15(11):e0242809. doi: 10.1371/journal.pone.0242809. eCollection 2020. PLoS One. 2020. PMID: 33253299 Free PMC article.
-
Aldehyde Dehydrogenase Enzyme Functions in Acute Leukemia Stem Cells.Front Biosci (Schol Ed). 2022 Mar 8;14(1):8. doi: 10.31083/j.fbs1401008. Front Biosci (Schol Ed). 2022. PMID: 35320919 Review.
Cited by
-
OGG1 as an Epigenetic Reader Affects NFκB: What This Means for Cancer.Cancers (Basel). 2023 Dec 28;16(1):148. doi: 10.3390/cancers16010148. Cancers (Basel). 2023. PMID: 38201575 Free PMC article. Review.
-
Investigating the biology of microRNA links to ALDH1A1 reveals candidates for preclinical testing in acute myeloid leukemia.Int J Oncol. 2024 Dec;65(6):115. doi: 10.3892/ijo.2024.5703. Epub 2024 Nov 8. Int J Oncol. 2024. PMID: 39513593 Free PMC article. Review.
-
Divergent Processing of Cell Stress Signals as the Basis of Cancer Progression: Licensing NFκB on Chromatin.Int J Mol Sci. 2024 Aug 7;25(16):8621. doi: 10.3390/ijms25168621. Int J Mol Sci. 2024. PMID: 39201306 Free PMC article. Review.
-
Epithelial-Mesenchymal Transition in Acute Leukemias.Int J Mol Sci. 2024 Feb 11;25(4):2173. doi: 10.3390/ijms25042173. Int J Mol Sci. 2024. PMID: 38396852 Free PMC article. Review.
References
-
- Moreb J., Schweder M., Suresh A., Zucali J.R. Overexpression of the Human Aldehyde Dehydrogenase Class I Results in Increased Resistance to 4-Hydroperoxycyclophosphamide. Cancer Gene Ther. 1996;3:24–30. - PubMed
-
- Vlahopoulos S.A., Cen O., Hengen N., Agan J., Moschovi M., Critselis E., Adamaki M., Bacopoulou F., Copland J.A., Boldogh I., et al. Dynamic Aberrant NF-ΚB Spurs Tumorigenesis: A New Model Encompassing the Microenvironment. Cytokine Growth Factor Rev. 2015;26:389–403. doi: 10.1016/j.cytogfr.2015.06.001. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

