Canine Somatic Mutations from Whole-Exome Sequencing of B-Cell Lymphomas in Six Canine Breeds-A Preliminary Study
- PMID: 37760246
- PMCID: PMC10525272
- DOI: 10.3390/ani13182846
Canine Somatic Mutations from Whole-Exome Sequencing of B-Cell Lymphomas in Six Canine Breeds-A Preliminary Study
Abstract
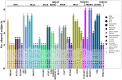
Canine lymphoma (CL) is one of the most common malignant tumors in dogs. The cause of CL remains unclear. Genetic mutations that have been suggested as possible causes of CL are not fully understood. Whole-exome sequencing (WES) is a time- and cost-effective method for detecting genetic variants targeting only the protein-coding regions (exons) that are part of the entire genome region. A total of eight patients with B-cell lymphomas were recruited, and WES analysis was performed on whole blood and lymph node aspirate samples from each patient. A total of 17 somatic variants (GOLIM4, ITM2B, STN1, UNC79, PLEKHG4, BRF1, ENSCAFG00845007156, SEMA6B, DSC1, TNFAIP1, MYLK3, WAPL, ADORA2B, LOXHD1, GP6, AZIN1, and NCSTN) with moderate to high impact were identified by WES analysis. Through a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of 17 genes with somatic mutations, a total of 16 pathways were identified. Overall, the somatic mutations identified in this study suggest novel candidate mutations for CL, and further studies are needed to confirm the role of these mutations.
Keywords: B-cell; PARR; canine lymphoma; whole-exome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations.Hum Reprod. 2016 Dec;31(12):2872-2880. doi: 10.1093/humrep/dew262. Epub 2016 Oct 26. Hum Reprod. 2016. PMID: 27798045
-
Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background.Genome Res. 2015 Nov;25(11):1634-45. doi: 10.1101/gr.194449.115. Epub 2015 Sep 16. Genome Res. 2015. PMID: 26377837 Free PMC article.
-
Detection and benchmarking of somatic mutations in cancer genomes using RNA-seq data.PeerJ. 2018 Jul 31;6:e5362. doi: 10.7717/peerj.5362. eCollection 2018. PeerJ. 2018. PMID: 30083469 Free PMC article.
-
Next generation sequencing and the management of diffuse large B-cell lymphoma: from whole exome analysis to targeted therapy.Discov Med. 2014 Jul-Aug;18(97):51-65. Discov Med. 2014. PMID: 25091488 Review.
-
Activating somatic mutations in diffuse large B-cell lymphomas: lessons from next generation sequencing and key elements in the precision medicine era.Leuk Lymphoma. 2015 May;56(5):1213-22. doi: 10.3109/10428194.2014.941836. Epub 2014 Oct 9. Leuk Lymphoma. 2015. PMID: 25130477 Review.
References
-
- Sastre L. Exome Sequencing: What Clinicians Need to Know. Adv. Genom. Genet. 2014;4:15–27. doi: 10.2147/AGG.S39108. - DOI
-
- Kunstman J.W., Juhlin C.C., Goh G., Brown T.C., Stenman A., Healy J.M., Rubinstein J.C., Choi M., Kiss N., Nelson-Williams C., et al. Characterization of the Mutational Landscape of Anaplastic Thyroid Cancer via Whole-Exome Sequencing. Hum. Mol. Genet. 2015;24:2318–2329. doi: 10.1093/hmg/ddu749. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

