Inhibition of fatty acid oxidation enables heart regeneration in adult mice
- PMID: 37758950
- PMCID: PMC10584682
- DOI: 10.1038/s41586-023-06585-5
Inhibition of fatty acid oxidation enables heart regeneration in adult mice
Erratum in
-
Publisher Correction: Inhibition of fatty acid oxidation enables heart regeneration in adult mice.Nature. 2023 Nov;623(7986):E7. doi: 10.1038/s41586-023-06755-5. Nature. 2023. PMID: 37863963 Free PMC article. No abstract available.
Abstract
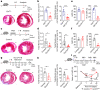
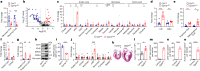
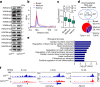
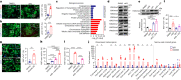
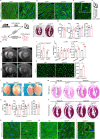
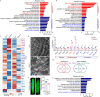
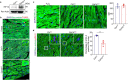
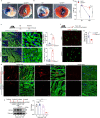
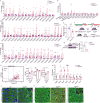
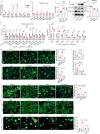
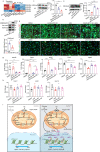
Postnatal maturation of cardiomyocytes is characterized by a metabolic switch from glycolysis to fatty acid oxidation, chromatin reconfiguration and exit from the cell cycle, instating a barrier for adult heart regeneration1,2. Here, to explore whether metabolic reprogramming can overcome this barrier and enable heart regeneration, we abrogate fatty acid oxidation in cardiomyocytes by inactivation of Cpt1b. We find that disablement of fatty acid oxidation in cardiomyocytes improves resistance to hypoxia and stimulates cardiomyocyte proliferation, allowing heart regeneration after ischaemia-reperfusion injury. Metabolic studies reveal profound changes in energy metabolism and accumulation of α-ketoglutarate in Cpt1b-mutant cardiomyocytes, leading to activation of the α-ketoglutarate-dependent lysine demethylase KDM5 (ref. 3). Activated KDM5 demethylates broad H3K4me3 domains in genes that drive cardiomyocyte maturation, lowering their transcription levels and shifting cardiomyocytes into a less mature state, thereby promoting proliferation. We conclude that metabolic maturation shapes the epigenetic landscape of cardiomyocytes, creating a roadblock for further cell divisions. Reversal of this process allows repair of damaged hearts.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Metabolic reprogramming unlocks the regenerative potential of the heart.Nat Rev Cardiol. 2023 Dec;20(12):795. doi: 10.1038/s41569-023-00945-4. Nat Rev Cardiol. 2023. PMID: 37833501 No abstract available.
-
Fueling cardiac myocyte proliferation.J Cardiovasc Aging. 2024 Jan;4(1):2. doi: 10.20517/jca.2023.47. Epub 2023 Dec 31. J Cardiovasc Aging. 2024. PMID: 38455511 Free PMC article.
Similar articles
-
PHDs/CPT1B/VDAC1 axis regulates long-chain fatty acid oxidation in cardiomyocytes.Cell Rep. 2021 Oct 5;37(1):109767. doi: 10.1016/j.celrep.2021.109767. Cell Rep. 2021. PMID: 34610308 Free PMC article.
-
Histone demethylase KDM5 regulates cardiomyocyte maturation by promoting fatty acid oxidation, oxidative phosphorylation, and myofibrillar organization.Cardiovasc Res. 2024 May 7;120(6):630-643. doi: 10.1093/cvr/cvae014. Cardiovasc Res. 2024. PMID: 38230606
-
RNA-Binding Protein LIN28a Regulates New Myocyte Formation in the Heart Through Long Noncoding RNA-H19.Circulation. 2023 Jan 24;147(4):324-337. doi: 10.1161/CIRCULATIONAHA.122.059346. Epub 2022 Oct 31. Circulation. 2023. PMID: 36314132 Free PMC article.
-
Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation.J Cardiovasc Pharmacol. 2010 Aug;56(2):130-40. doi: 10.1097/FJC.0b013e3181e74a14. J Cardiovasc Pharmacol. 2010. PMID: 20505524 Review.
-
Metabolic Reprogramming: A Byproduct or a Driver of Cardiomyocyte Proliferation?Circulation. 2024 May 14;149(20):1598-1610. doi: 10.1161/CIRCULATIONAHA.123.065880. Epub 2024 May 13. Circulation. 2024. PMID: 38739695 Review.
Cited by
-
Carnitine is a friend in HFpEF and foe in HFrEF.iScience. 2024 Sep 23;27(10):111018. doi: 10.1016/j.isci.2024.111018. eCollection 2024 Oct 18. iScience. 2024. PMID: 39429785 Free PMC article.
-
Intermedin Alleviates Diabetic Cardiomyopathy by Up-Regulating CPT-1β through Activation of the Phosphatidyl Inositol 3 Kinase/Protein Kinase B Signaling Pathway.Pharmaceuticals (Basel). 2024 Sep 12;17(9):1204. doi: 10.3390/ph17091204. Pharmaceuticals (Basel). 2024. PMID: 39338366 Free PMC article.
-
Maternal high-fat diet alters Tet-mediated epigenetic regulation during heart development.iScience. 2024 Jul 31;27(9):110631. doi: 10.1016/j.isci.2024.110631. eCollection 2024 Sep 20. iScience. 2024. PMID: 39262804 Free PMC article.
-
Chemically programmed metabolism drives a superior cell fitness for cartilage regeneration.Sci Adv. 2024 Sep 13;10(37):eadp4408. doi: 10.1126/sciadv.adp4408. Epub 2024 Sep 11. Sci Adv. 2024. PMID: 39259800 Free PMC article.
-
α-Ketoglutarate promotes cardiomyocyte proliferation and heart regeneration after myocardial infarction.Nat Cardiovasc Res. 2024 Sep;3(9):1083-1097. doi: 10.1038/s44161-024-00531-y. Epub 2024 Sep 2. Nat Cardiovasc Res. 2024. PMID: 39223390
References
-
- Nakada, Y. et al. Hypoxia induces heart regeneration in adult mice. Nature10.1038/nature20173 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

