Activated Src kinase promotes cell cannibalism in Drosophila
- PMID: 37747450
- PMCID: PMC10518265
- DOI: 10.1083/jcb.202302076
Activated Src kinase promotes cell cannibalism in Drosophila
Abstract
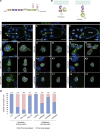
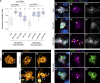
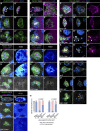
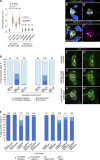
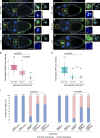
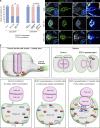
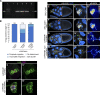
Src family kinases (SFKs) are evolutionarily conserved proteins acting downstream of receptors and regulating cellular processes including proliferation, adhesion, and migration. Elevated SFK expression and activity correlate with progression of a variety of cancers. Here, using the Drosophila melanogaster border cells as a model, we report that localized activation of a Src kinase promotes an unusual behavior: engulfment of one cell by another. By modulating Src expression and activity in the border cell cluster, we found that increased Src kinase activity, either by mutation or loss of a negative regulator, is sufficient to drive one cell to engulf another living cell. We elucidate a molecular mechanism that requires integrins, the kinases SHARK and FAK, and Rho family GTPases, but not the engulfment receptor Draper. We propose that cell cannibalism is a result of aberrant phagocytosis, where cells with dysregulated Src activity fail to differentiate between living and dead or self versus non-self, thus driving this malignant behavior.
© 2023 Torres et al.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. A.Y. Torres reported a patent to GENETICALLY ENGINEERED PHAGOCYTES, AND RELATED COMPOSITIONS, VECTORS, METHODS, AND SYSTEMS pending. D.J. Montell reported other from Inceptor Bio outside the submitted work; in addition, D.J. Montell had a patent to 17/995,083 pending “Inceptor Bio.” No other disclosures were reported.
Figures

Similar articles
-
Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling.Nature. 2008 Jun 12;453(7197):935-9. doi: 10.1038/nature06901. Epub 2008 Apr 23. Nature. 2008. PMID: 18432193 Free PMC article.
-
SRC catalytic but not scaffolding function is needed for integrin-regulated tyrosine phosphorylation, cell migration, and cell spreading.Mol Cell Biol. 2002 Apr;22(8):2427-40. doi: 10.1128/MCB.22.8.2427-2440.2002. Mol Cell Biol. 2002. PMID: 11909938 Free PMC article.
-
Src family kinases are required for integrin but not PDGFR signal transduction.EMBO J. 1999 May 4;18(9):2459-71. doi: 10.1093/emboj/18.9.2459. EMBO J. 1999. PMID: 10228160 Free PMC article.
-
Src kinases as therapeutic targets for cancer.Nat Rev Clin Oncol. 2009 Oct;6(10):587-95. doi: 10.1038/nrclinonc.2009.129. Nat Rev Clin Oncol. 2009. PMID: 19787002 Review.
-
[SRC kinases in tumor therapy].Med Klin (Munich). 2010 Oct;105(10):711-5. doi: 10.1007/s00063-010-1123-y. Epub 2010 Oct 28. Med Klin (Munich). 2010. PMID: 20981590 Review. German.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

