IGF2BP3 mediates the mRNA degradation of NF1 to promote triple-negative breast cancer progression via an m6A-dependent manner
- PMID: 37743642
- PMCID: PMC10518495
- DOI: 10.1002/ctm2.1427
IGF2BP3 mediates the mRNA degradation of NF1 to promote triple-negative breast cancer progression via an m6A-dependent manner
Abstract
Background: N6-methyladenosine (m6A) is an abundant reversible modification in eukaryotic mRNAs. Emerging evidences indicate that m6A modification plays a vital role in tumourigenesis. As a crucial reader of m6A, IGF2BP3 usually mediates the stabilisation of mRNAs via an m6A-dependent manner. But the underlying mechanism of IGF2BP3 in the tumourigenesis of triple-negative breast cancer (TNBC) is unclear.
Methods: TCGA cohorts were analysed for IGF2BP3 expression and IGF2BP3 promoter methylation levels in different breast cancer subtypes. Colony formation, flow cytometry assays and subcutaneous xenograft were performed to identify the phenotype of IGF2BP3 in TNBC. RNA/RNA immunoprecipitation (RIP)/methylated RNA immunoprecipitation (MeRIP) sequencing and luciferase assays were used to certify the target of IGF2BP3 in TNBC cells.
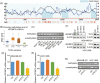
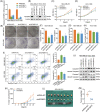
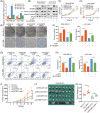
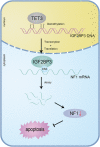
Results: IGF2BP3 was highly expressed in TNBC cell lines and tissues. TET3-mediated IGF2BP3 promoter hypomethylation led to the upregulation of IGF2BP3. Knocking down IGF2BP3 markedly reduced the proliferation of TNBC in vitro and in vivo. Intersection co-assays revealed that IGF2BP3 decreased neurofibromin 1 (NF1) stabilisation via an m6A-dependent manner. NF1 knockdown could rescue the phenotypes of IGF2BP3 knockdown cells partially.
Conclusion: TET3-mediated IGF2BP3 accelerated the proliferation of TNBC by destabilising NF1 mRNA via an m6A-dependent manner. This suggests that IGF2BP3 could be a potential therapeutic target for TNBC.
Keywords: IGF2BP3; NF1; TET3; TNBC; m6A.
© 2023 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures




Similar articles
-
circBRAF promotes the progression of triple-negative breast cancer through modulating methylation by recruiting KDM4B to histone H3K9me3 and IGF2BP3 to mRNA.Am J Cancer Res. 2024 May 15;14(5):2020-2036. doi: 10.62347/OOLG5765. eCollection 2024. Am J Cancer Res. 2024. PMID: 38859856 Free PMC article.
-
RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer.J Exp Clin Cancer Res. 2020 Sep 29;39(1):203. doi: 10.1186/s13046-020-01714-8. J Exp Clin Cancer Res. 2020. PMID: 32993738 Free PMC article.
-
RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent.J Exp Clin Cancer Res. 2021 Feb 26;40(1):80. doi: 10.1186/s13046-021-01871-4. J Exp Clin Cancer Res. 2021. PMID: 33637103 Free PMC article.
-
RNA m6A reader IGF2BP3 promotes metastasis of triple-negative breast cancer via SLIT2 repression.FASEB J. 2022 Nov;36(11):e22618. doi: 10.1096/fj.202200751RR. FASEB J. 2022. PMID: 36250924
-
Targeting IGF2BP3 in Cancer.Int J Mol Sci. 2023 May 29;24(11):9423. doi: 10.3390/ijms24119423. Int J Mol Sci. 2023. PMID: 37298373 Free PMC article. Review.
Cited by
-
circBRAF promotes the progression of triple-negative breast cancer through modulating methylation by recruiting KDM4B to histone H3K9me3 and IGF2BP3 to mRNA.Am J Cancer Res. 2024 May 15;14(5):2020-2036. doi: 10.62347/OOLG5765. eCollection 2024. Am J Cancer Res. 2024. PMID: 38859856 Free PMC article.
-
Dissecting the effects of METTL3 on alternative splicing in prostate cancer.Front Oncol. 2023 Aug 22;13:1227016. doi: 10.3389/fonc.2023.1227016. eCollection 2023. Front Oncol. 2023. PMID: 37675218 Free PMC article.
-
LRPPRC promotes glycolysis by stabilising LDHA mRNA and its knockdown plus glutamine inhibitor induces synthetic lethality via m6 A modification in triple-negative breast cancer.Clin Transl Med. 2024 Feb;14(2):e1583. doi: 10.1002/ctm2.1583. Clin Transl Med. 2024. PMID: 38372449 Free PMC article.
-
NRF2 activation by cysteine as a survival mechanism for triple-negative breast cancer cells.Oncogene. 2024 May;43(22):1701-1713. doi: 10.1038/s41388-024-03025-0. Epub 2024 Apr 10. Oncogene. 2024. PMID: 38600165 Free PMC article.
-
Berberine inhibits the progression of breast cancer by regulating METTL3-mediated m6A modification of FGF7 mRNA.Thorac Cancer. 2024 Jun;15(17):1357-1368. doi: 10.1111/1759-7714.15321. Epub 2024 May 6. Thorac Cancer. 2024. PMID: 38709912 Free PMC article.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7‐33. - PubMed
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. - PubMed
-
- Lu H. New players critical for breast cancer. J Mol Cell Biol. 2018;10:271‐272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
