Generation of recombinant mAbs to vaccinia virus displaying high affinity and potent neutralization
- PMID: 37737634
- PMCID: PMC10581037
- DOI: 10.1128/spectrum.01598-23
Generation of recombinant mAbs to vaccinia virus displaying high affinity and potent neutralization
Abstract
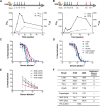
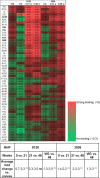
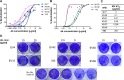
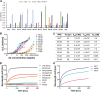
Members of the Orthopoxvirus genus can cause severe infections in humans. Global vaccination against smallpox, caused by the variola virus, resulted in the eradication of the disease in 1980. Shortly thereafter, vaccination was discontinued, and as a result, a large proportion of the current population is not protected against orthopoxviruses. The concerns that the variola virus or other engineered forms of poxviruses may re-emerge as bioweapons and the sporadic outbreaks of zoonotic members of the family, such as Mpox, which are becoming more frequent and prevalent, also emphasize the need for an effective treatment against orthopoxviruses. To date, the most effective way to prevent or control an orthopoxvirus outbreak is through vaccination. However, the traditional vaccinia-based vaccine may cause severe side effects. Vaccinia immune globulin was approved by the U.S. Food and Drug Administration (FDA) for the treatment of vaccine adverse reactions and was also used occasionally for the treatment of severe orthopoxvirus infections. However, this treatment carries many disadvantages and is also in short supply. Thus, a recombinant alternative is highly needed. In this study, two non-human primates were immunized with live vaccinia virus, producing a robust and diverse antibody response. A phage-display library was constructed based on the animal's lymphatic organs, and a panel of neutralizing monoclonal antibodies (mAbs), recognizing diverse proteins of the vaccinia virus, was selected and characterized. These antibodies recognized both mature virion and enveloped virion forms of the virus and exhibited high affinity and potent in vitro neutralization capabilities. Furthermore, these monoclonal antibodies were able to neutralize Mpox 2018 and 2022 strains, suggesting a potential for cross-species protection. We suggest that a combination of these mAbs has the potential to serve as recombinant therapy both for vaccinia vaccine adverse reactions and for orthopoxvirus infections. IMPORTANCE In this manuscript, we report the isolation and characterization of several recombinant neutralizing monoclonal antibodies (mAbs) identified by screening a phage-display library constructed from lymphatic cells collected from immunized non-human primates. The antibodies target several different antigens of the vaccinia virus, covering both mature virion and extracellular enveloped virion forms of the virus. We document strong evidence indicating that they exhibit excellent affinity to their respective antigens and, most importantly, optimal in vitro neutralization of the virus, which exceeded that of vaccinia immune globulin. Furthermore, we present the ability of these novel isolated mAbs (as well as the sera collected from vaccinia-immunized animals) to neutralize two Mpox strains from the 2018 to 2022 outbreaks. We believe that these antibodies have the potential to be used for the treatment of vaccinia vaccine adverse reactions, for other orthopoxvirus infections, and in cases of unexpected bioterror scenarios.
Keywords: Mpox; VIG; neutralizing antibodies; orthopoxviruses; vaccinia virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Neutralization Determinants on Poxviruses.Viruses. 2023 Dec 8;15(12):2396. doi: 10.3390/v15122396. Viruses. 2023. PMID: 38140637 Free PMC article. Review.
-
Potent neutralization of vaccinia virus by divergent murine antibodies targeting a common site of vulnerability in L1 protein.J Virol. 2014 Oct;88(19):11339-55. doi: 10.1128/JVI.01491-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031354 Free PMC article.
-
Cross-Neutralizing and Protective Human Antibody Specificities to Poxvirus Infections.Cell. 2016 Oct 20;167(3):684-694.e9. doi: 10.1016/j.cell.2016.09.049. Cell. 2016. PMID: 27768891 Free PMC article.
-
Selection of Vaccinia Virus-Neutralizing Antibody from a Phage-Display Human-Antibody Library.J Microbiol Biotechnol. 2019 Apr 28;29(4):651-657. doi: 10.4014/jmb.1812.12024. J Microbiol Biotechnol. 2019. PMID: 30856707
-
The Global Monkeypox (Mpox) Outbreak: A Comprehensive Review.Vaccines (Basel). 2023 Jun 12;11(6):1093. doi: 10.3390/vaccines11061093. Vaccines (Basel). 2023. PMID: 37376482 Free PMC article. Review.
Cited by
-
Synergistic effect of two human-like monoclonal antibodies confers protection against orthopoxvirus infection.Nat Commun. 2024 Apr 16;15(1):3265. doi: 10.1038/s41467-024-47328-y. Nat Commun. 2024. PMID: 38627363 Free PMC article.
-
Neutralization Determinants on Poxviruses.Viruses. 2023 Dec 8;15(12):2396. doi: 10.3390/v15122396. Viruses. 2023. PMID: 38140637 Free PMC article. Review.
References
-
- José da Silva Domingos I, Silva de Oliveira J, Lorene Soares Rocha K, Bretas de Oliveira D, Geessien Kroon E, Barbosa Costa G, de Souza Trindade G. 2021. Twenty years after bovine vaccinia in Brazil: where we are and where are we going. Pathogens 10:406. doi:10.3390/pathogens10040406 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

