MAST4 promotes primary ciliary resorption through phosphorylation of Tctex-1
- PMID: 37726137
- PMCID: PMC10509483
- DOI: 10.26508/lsa.202301947
MAST4 promotes primary ciliary resorption through phosphorylation of Tctex-1
Abstract
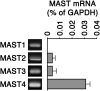
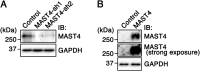
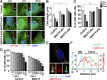
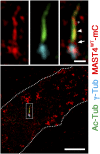
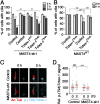
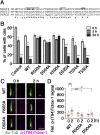
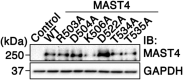
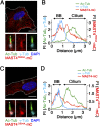
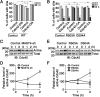
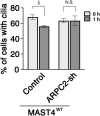
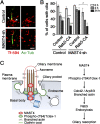
The primary cilium undergoes cell cycle-dependent assembly and disassembly. Dysregulated ciliary dynamics are associated with several pathological conditions called ciliopathies. Previous studies showed that the localization of phosphorylated Tctex-1 at Thr94 (T94) at the ciliary base critically regulates ciliary resorption by accelerating actin remodeling and ciliary pocket membrane endocytosis. Here, we show that microtubule-associated serine/threonine kinase family member 4 (MAST4) is localized at the primary cilium. Suppressing MAST4 blocks serum-induced ciliary resorption, and overexpressing MAST4 accelerates ciliary resorption. Tctex-1 binds to the kinase domain of MAST4, in which the R503 and D504 residues are key to MAST4-mediated ciliary resorption. The ciliary resorption and the ciliary base localization of phospho-(T94)Tctex-1 are blocked by the knockdown of MAST4 or the expression of the catalytic-inactive site-directed MAST4 mutants. Moreover, MAST4 is required for Cdc42 activation and Rab5-mediated periciliary membrane endocytosis during ciliary resorption. These results support that MAST4 is a novel kinase that regulates ciliary resorption by modulating the ciliary base localization of phospho-(T94)Tctex-1. MAST4 is a potential new target for treating ciliopathies causally by ciliary resorption defects.
© 2023 Sakaji et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7 PMID: 38260445 Free PMC article. Updated. Preprint.
-
Akt1 sequentially phosphorylates p27kip1 within a conserved but non-canonical region.Cell Div. 2006 Jun 16;1:11. doi: 10.1186/1747-1028-1-11. Cell Div. 2006. PMID: 16780593 Free PMC article.
-
The effectiveness of school-based family asthma educational programs on the quality of life and number of asthma exacerbations of children aged five to 18 years diagnosed with asthma: a systematic review protocol.JBI Database System Rev Implement Rep. 2015 Oct;13(10):69-81. doi: 10.11124/jbisrir-2015-2335. JBI Database System Rev Implement Rep. 2015. PMID: 26571284
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Platelet-rich therapies for musculoskeletal soft tissue injuries.Cochrane Database Syst Rev. 2014 Apr 29;2014(4):CD010071. doi: 10.1002/14651858.CD010071.pub3. Cochrane Database Syst Rev. 2014. PMID: 24782334 Free PMC article. Review.
Cited by
-
Patient-derived and gene-edited pluripotent stem cells lacking NPHP1 recapitulate juvenile nephronophthisis in abnormalities of primary cilia and renal cyst formation.Front Cell Dev Biol. 2024 Jun 26;12:1370723. doi: 10.3389/fcell.2024.1370723. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38989059 Free PMC article.
References
-
- Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MP, Pedersen LB, Benmerah A, Andersen CY, Larsen LA, et al. (2013) TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep 3: 1806–1814. 10.1016/j.celrep.2013.05.020 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
