Spontaneous HIV expression during suppressive ART is associated with the magnitude and function of HIV-specific CD4+ and CD8+ T cells
- PMID: 37708853
- PMCID: PMC10542967
- DOI: 10.1016/j.chom.2023.08.006
Spontaneous HIV expression during suppressive ART is associated with the magnitude and function of HIV-specific CD4+ and CD8+ T cells
Abstract
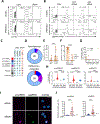
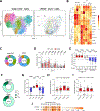
Spontaneous transcription and translation of HIV can persist during suppressive antiretroviral therapy (ART). The quantity, phenotype, and biological relevance of this spontaneously "active" reservoir remain unclear. Using multiplexed single-cell RNAflow-fluorescence in situ hybridization (FISH), we detect active HIV transcription in 14/18 people with HIV on suppressive ART, with a median of 28/million CD4+ T cells. While these cells predominantly exhibit abortive transcription, p24-expressing cells are evident in 39% of participants. Phenotypically diverse, active reservoirs are enriched in central memory T cells and CCR6- and activation-marker-expressing cells. The magnitude of the active reservoir positively correlates with total HIV-specific CD4+ and CD8+ T cell responses and with multiple HIV-specific T cell clusters identified by unsupervised analysis. These associations are particularly strong with p24-expressing active reservoir cells. Single-cell vDNA sequencing shows that active reservoirs are largely dominated by defective proviruses. Our data suggest that these reservoirs maintain HIV-specific CD4+ and CD8+ T responses during suppressive ART.
Keywords: HIV; HIV-specific CD4 T cell responses; HIV-specific CD8 T cell responses; antiretroviral therapy; flow cytometric fluorescence in situ RNA hybridization; spontaneously active reservoirs; viral reservoirs; viral transcription; viral translation.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
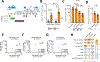



Similar articles
-
Combined single-cell transcriptional, translational, and genomic profiling reveals HIV-1 reservoir diversity.Cell Rep. 2021 Aug 31;36(9):109643. doi: 10.1016/j.celrep.2021.109643. Cell Rep. 2021. PMID: 34469719
-
An active HIV reservoir during ART is associated with maintenance of HIV-specific CD8+ T cell magnitude and short-lived differentiation status.Cell Host Microbe. 2023 Sep 13;31(9):1494-1506.e4. doi: 10.1016/j.chom.2023.08.012. Cell Host Microbe. 2023. PMID: 37708852 Free PMC article.
-
A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4+ T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients.mBio. 2017 Jul 11;8(4):e00876-17. doi: 10.1128/mBio.00876-17. mBio. 2017. PMID: 28698276 Free PMC article.
-
Persistent HIV-1 transcription during ART: time to reassess its significance?Curr Opin HIV AIDS. 2024 May 1;19(3):124-132. doi: 10.1097/COH.0000000000000849. Epub 2024 Mar 12. Curr Opin HIV AIDS. 2024. PMID: 38502547 Free PMC article. Review.
-
Assessing proviral competence: current approaches to evaluate HIV-1 persistence.Curr Opin HIV AIDS. 2021 Jul 1;16(4):223-231. doi: 10.1097/COH.0000000000000687. Curr Opin HIV AIDS. 2021. PMID: 33993171 Review.
Cited by
-
Mechanisms and efficacy of small molecule latency-promoting agents to inhibit HIV reactivation ex vivo.JCI Insight. 2024 Aug 20;9(19):e183084. doi: 10.1172/jci.insight.183084. JCI Insight. 2024. PMID: 39163135 Free PMC article.
-
Progress Note 2024: Curing HIV; Not in My Lifetime or Just Around the Corner?Pathog Immun. 2024 Mar 1;8(2):115-157. doi: 10.20411/pai.v8i2.665. eCollection 2023. Pathog Immun. 2024. PMID: 38455668 Free PMC article.
-
Distinguishable topology of the task-evoked functional genome networks in HIV-1 reservoirs.iScience. 2024 Oct 21;27(11):111222. doi: 10.1016/j.isci.2024.111222. eCollection 2024 Nov 15. iScience. 2024. PMID: 39559761 Free PMC article.
-
Centrosome amplification and aneuploidy driven by the HIV-1-induced Vpr•VprBP•Plk4 complex in CD4+ T cells.Nat Commun. 2024 Mar 5;15(1):2017. doi: 10.1038/s41467-024-46306-8. Nat Commun. 2024. PMID: 38443376 Free PMC article.
-
Clonal succession after prolonged antiretroviral therapy rejuvenates CD8+ T cell responses against HIV-1.Nat Immunol. 2024 Sep;25(9):1555-1564. doi: 10.1038/s41590-024-01931-9. Epub 2024 Aug 23. Nat Immunol. 2024. PMID: 39179934
References
-
- Davey RT Jr., Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, et al. (1999). HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proceedings of the National Academy of Sciences of the United States of America 96, 15109–15114. 10.1073/pnas.96.26.15109. - DOI - PMC - PubMed
-
- Gaebler C, Lorenzi JCC, Oliveira TY, Nogueira L, Ramos V, Lu CL, Pai JA, Mendoza P, Jankovic M, Caskey M, and Nussenzweig MC (2019). Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. The Journal of experimental medicine 216, 2253–2264. 10.1084/jem.20190896. - DOI - PMC - PubMed
-
- Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, Richman DD, O’Doherty U, Palmer S, Hecht FM, et al. (2015). A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine 2, 874–883. 10.1016/j.ebiom.2015.06.019. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

