NK cell expansion requires HuR and mediates control of solid tumors and long-term virus infection
- PMID: 37698554
- PMCID: PMC10497399
- DOI: 10.1084/jem.20231154
NK cell expansion requires HuR and mediates control of solid tumors and long-term virus infection
Abstract
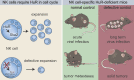
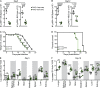
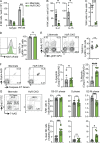
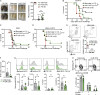
Natural killer (NK) cells are lymphocytes capable of controlling tumors and virus infections through direct lysis and cytokine production. While both T and NK cells expand and accumulate in affected tissues, the role of NK cell expansion in tumor and viral control is not well understood. Here, we show that posttranscriptional regulation by the RNA-binding protein HuR is essential for NK cell expansion without negatively affecting effector functions. HuR-deficient NK cells displayed defects in the metaphase of the cell cycle, including decreased expression and alternative splicing of Ska2, a component of the spindle and kinetochore complex. HuR-dependent NK cell expansion contributed to long-term cytomegalovirus control and facilitated control of subcutaneous tumors but not tumor metastases in two independent tumor models. These results show that posttranscriptional regulation by HuR specifically affects NK cell expansion, which is required for the control of long-term virus infection and solid tumors, but not acute infection or tumor metastases, highlighting fundamental differences with antigen-specific T cell control.
© 2023 Piersma et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







Similar articles
-
Leukocyte Immunoglobulin-Like Receptor 1-Expressing Human Natural Killer Cell Subsets Differentially Recognize Isolates of Human Cytomegalovirus through the Viral Major Histocompatibility Complex Class I Homolog UL18.J Virol. 2016 Jan 6;90(6):3123-37. doi: 10.1128/JVI.02614-15. J Virol. 2016. PMID: 26739048 Free PMC article.
-
Cytoplasmic overexpression of RNA-binding protein HuR is a marker of poor prognosis in meningioma, and HuR knockdown decreases meningioma cell growth and resistance to hypoxia.J Pathol. 2017 Aug;242(4):421-434. doi: 10.1002/path.4916. Epub 2017 Jul 5. J Pathol. 2017. PMID: 28493484
-
Natural killer cell effector functions in antiviral defense.FEBS J. 2022 Jul;289(14):3982-3999. doi: 10.1111/febs.16073. Epub 2021 Jun 30. FEBS J. 2022. PMID: 34125493 Review.
-
RNA-Binding Protein HuR Regulates Paneth Cell Function by Altering Membrane Localization of TLR2 via Post-transcriptional Control of CNPY3.Gastroenterology. 2019 Sep;157(3):731-743. doi: 10.1053/j.gastro.2019.05.010. Epub 2019 May 17. Gastroenterology. 2019. PMID: 31103627 Free PMC article.
-
The RNA-Binding Protein HuR in Digestive System Tumors.Biomed Res Int. 2020 Jul 24;2020:9656051. doi: 10.1155/2020/9656051. eCollection 2020. Biomed Res Int. 2020. PMID: 32775456 Free PMC article. Review.
Cited by
-
Cuproptosis-Related Genes as Prognostic Biomarkers for Sepsis: Insights into Immune Function and Personalized Immunotherapy.J Inflamm Res. 2024 Jul 2;17:4229-4245. doi: 10.2147/JIR.S461766. eCollection 2024. J Inflamm Res. 2024. PMID: 38979432 Free PMC article.
-
Tissue-specific features of innate lymphoid cells in antiviral defense.Cell Mol Immunol. 2024 Sep;21(9):1036-1050. doi: 10.1038/s41423-024-01161-x. Epub 2024 Apr 29. Cell Mol Immunol. 2024. PMID: 38684766 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

