Unraveling the role of ZNF506 as a human PBS-pro-targeting protein for ERVP repression
- PMID: 37697430
- PMCID: PMC10602909
- DOI: 10.1093/nar/gkad731
Unraveling the role of ZNF506 as a human PBS-pro-targeting protein for ERVP repression
Abstract
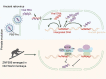
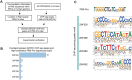
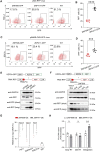
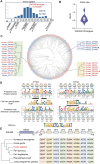
Krüppel-associated box zinc finger proteins (KZFPs) function as a defense mechanism to maintain the genome stability of higher vertebrates by regulating the transcriptional activities of transposable elements (TEs). While previous studies have characterized ZFP809 as responsible for binding and repressing ERVs containing a proline tRNA primer-binding site (PBS-Pro) in mice, comparable KZFPs have not been identified in humans yet. Here, we identified ZNF506 as a PBS-Pro-binding protein in humans, which functions as a transcriptional repressor of PBS-Pro-utilizing retroviruses by recruiting heterochromatic modifications. Although they have similar functions, the low protein similarities between ZNF506, ZFP809 and KZFPs of other species suggest their independent evolution against the invasion of PBS-Pro-utilizing retroviruses into their respective ancestor genomes after species divergence. We also explored the link between ZNF506 and leukemia. Our findings suggest that ZNF506 is a unique human KZFP that can bind to PBS-Pro, highlighting the diverse evolution of KZFPs in defending against retroviral invasions. Additionally, our study provides insights into the potential role of ZNF506 in leukemia, contributing to the expanding knowledge of KZFPs' crucial function in disease and genome stability.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks.Nature. 2017 Mar 23;543(7646):550-554. doi: 10.1038/nature21683. Epub 2017 Mar 8. Nature. 2017. PMID: 28273063
-
The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification.EMBO J. 2019 Sep 16;38(18):e101220. doi: 10.15252/embj.2018101220. Epub 2019 Aug 12. EMBO J. 2019. PMID: 31403225 Free PMC article.
-
The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses.Genes Dev. 2015 Mar 1;29(5):538-54. doi: 10.1101/gad.252767.114. Genes Dev. 2015. PMID: 25737282 Free PMC article.
-
Endogenous Retroviruses Walk a Fine Line between Priming and Silencing.Viruses. 2020 Jul 23;12(8):792. doi: 10.3390/v12080792. Viruses. 2020. PMID: 32718022 Free PMC article. Review.
-
Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview.Viruses. 2020 Aug 13;12(8):884. doi: 10.3390/v12080884. Viruses. 2020. PMID: 32823517 Free PMC article. Review.
References
-
- Friedman J.R., Fredericks W.J., Jensen D.E., Speicher D.W., Huang X.P., Neilson E.G., Rauscher F.J. 3rd. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996; 10:2067–2078. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

