Epstein-Barr virus-encoded miR-BART11-3p modulates the DUSP6-MAPK axis to promote gastric cancer cell proliferation and metastasis
- PMID: 37681959
- PMCID: PMC10537804
- DOI: 10.1128/jvi.00881-23
Epstein-Barr virus-encoded miR-BART11-3p modulates the DUSP6-MAPK axis to promote gastric cancer cell proliferation and metastasis
Abstract
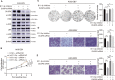
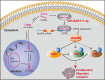
Epstein-Barr virus (EBV)-encoded miRNAs within the BamHI-A rightward transcript (BART) region are abundantly expressed in EBV-associated gastric cancer (EBVaGC), suggesting that they play roles in tumorigenesis. However, how these viral miRNAs contribute to the development of EBVaGC remains largely obscure. In this study, we found that EBV-encoded miR-BART11-3p targets 3' -UTR of dual-specificity phosphatase 6 (DUSP6) mRNA to upregulate ERK phosphorylation and downregulate JNK and p38 phosphorylation. By doing so, miR-BART11-3p promotes gastric cancer (GC) cell proliferation, migration, and invasion in vitro, and facilitates tumor growth in vivo. Restoration of DUSP6 expression reverses the tumor-promoting activity of miR-BART11-3p in AGS GC cells. Consistently, knockdown of DUSP6 ablates the antitumor effects of miR-BART11-3p inhibitors in EBV-positive GC cells. Furthermore, blocking ERK phosphorylation with trametinib inhibited the proliferation, migration, and invasion of miR-BART11-3p-expressing AGS cells. Administration of a miR-BART11-3p antagomir reduced the growth of EBV-positive xenograft tumors. Together, these findings reveal a novel mechanism by which EBV dysregulates MAPK pathways through an EBV-encoded microRNA to promote the development and progression of EBVaGC, which may be harnessed to develop new therapeutics to treat EBVaGC. IMPORTANCE The Epstein-Barr virus (EBV) is the first human tumor virus found to encode miRNAs, which within the BART region have been detected abundantly in EBV-associated gastric cancer (EBVaGC) and play various roles in promoting tumorigenesis. In our study, we observed that EBV-miR-BART11-3p promotes cell proliferation and induces migration and invasion in GC. Interestingly, we showed that miR-BART11-3p upregulates p-ERK and downregulates p-JNK and p-p38 by directly targeting 3'-UTR of dual-specificity phosphatase 6 (DUSP6). Restoration of DUSP6 rescues the effects generated by miR-BART11-3p in GC cells, and blocking ERK phosphorylation with Trametinib augments JNK and p38 phosphorylation and inhibits the effects of miR-BART11-3p-expressing AGS cells, suggesting that miR-BART11-3p promotes cell proliferation, migration, and invasion by modulating DUSP6-MAPK axis in EBVaGC. The findings presented in this study provide new mechanisms into the tumorigenesis in EBVaGC and new avenues for the development of therapeutic strategies to combat EBVaGC targeting miR-BART11-3p or phospho-ERK.
Keywords: EBV-miR-BART11-3p; Epstein-Barr virus; dual-specificity phosphatase 6; gastric cancer; mitogen-activated protein kinases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Epstein-Barr virus miR-BART2-5p and miR-BART11-5p regulate cell proliferation, apoptosis, and migration by targeting RB and p21 in gastric carcinoma.J Med Virol. 2023 Jan;95(1):e28338. doi: 10.1002/jmv.28338. J Med Virol. 2023. PMID: 36418188
-
Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression.J Virol. 2018 Nov 12;92(23):e01022-18. doi: 10.1128/JVI.01022-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209170 Free PMC article.
-
Epstein-Barr virus-encoded miR-BART11 promotes tumor-associated macrophage-induced epithelial-mesenchymal transition via targeting FOXP1 in gastric cancer.Virology. 2020 Sep;548:6-16. doi: 10.1016/j.virol.2020.05.011. Epub 2020 Jun 2. Virology. 2020. PMID: 32530809
-
The Role of LMP1 in Epstein-Barr Virus-associated Gastric Cancer.Curr Cancer Drug Targets. 2024;24(2):127-141. doi: 10.2174/1568009623666230512153741. Curr Cancer Drug Targets. 2024. PMID: 37183458 Review.
-
The role of EBV-encoded miRNA in EBV-associated gastric cancer.Front Oncol. 2023 Jun 14;13:1204030. doi: 10.3389/fonc.2023.1204030. eCollection 2023. Front Oncol. 2023. PMID: 37388232 Free PMC article. Review.
Cited by
-
Unlocking the role of non-coding RNAs in prostate cancer progression: exploring the interplay with the Wnt signaling pathway.Front Pharmacol. 2023 Sep 27;14:1269233. doi: 10.3389/fphar.2023.1269233. eCollection 2023. Front Pharmacol. 2023. PMID: 37829301 Free PMC article. Review.
-
Stearoyl-CoA desaturase 1 is targeted by EBV-encoded miR-BART20-5p and regulates cell autophagy, proliferation, and migration in EBV-associated gastric cancer.Virus Genes. 2024 Oct;60(5):464-474. doi: 10.1007/s11262-024-02094-3. Epub 2024 Aug 3. Virus Genes. 2024. PMID: 39096336
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

