Roles of small RNAs in crop disease resistance
- PMID: 37676520
- PMCID: PMC10429495
- DOI: 10.1007/s44154-021-00005-2
Roles of small RNAs in crop disease resistance
Abstract
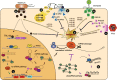
Small RNAs (sRNAs) are a class of short, non-coding regulatory RNAs that have emerged as critical components of defense regulatory networks across plant kingdoms. Many sRNA-based technologies, such as host-induced gene silencing (HIGS), spray-induced gene silencing (SIGS), virus-induced gene silencing (VIGS), artificial microRNA (amiRNA) and synthetic trans-acting siRNA (syn-tasiRNA)-mediated RNA interference (RNAi), have been developed as disease control strategies in both monocot and dicot plants, particularly in crops. This review aims to highlight our current understanding of the roles of sRNAs including miRNAs, heterochromatic siRNAs (hc-siRNAs), phased, secondary siRNAs (phasiRNAs) and natural antisense siRNAs (nat-siRNAs) in disease resistance, and sRNAs-mediated trade-offs between defense and growth in crops. In particular, we focus on the diverse functions of sRNAs in defense responses to bacterial and fungal pathogens, oomycete and virus in crops. Further, we highlight the application of sRNA-based technologies in protecting crops from pathogens. Further research perspectives are proposed to develop new sRNAs-based efficient strategies to breed non-genetically modified (GMO), disease-tolerant crops for sustainable agriculture.
Keywords: Crop diseases; HIGS; RNAi; RNAi-based technology; Small RNAs.
© 2021. The Author(s).
Conflict of interest statement
Author Z.H. is a member of the Editorial Board and was not involved in the journal's review of, or decisions related to, this manuscript.
Figures

Similar articles
-
Secondary siRNAs in Plants: Biosynthesis, Various Functions, and Applications in Virology.Front Plant Sci. 2021 Mar 2;12:610283. doi: 10.3389/fpls.2021.610283. eCollection 2021. Front Plant Sci. 2021. PMID: 33737942 Free PMC article. Review.
-
Systemic silencing of an endogenous plant gene by two classes of mobile 21-nucleotide artificial small RNAs.Plant J. 2022 May;110(4):1166-1181. doi: 10.1111/tpj.15730. Epub 2022 Mar 27. Plant J. 2022. PMID: 35277899 Free PMC article.
-
Cross-Kingdom Small RNAs Among Animals, Plants and Microbes.Cells. 2019 Apr 23;8(4):371. doi: 10.3390/cells8040371. Cells. 2019. PMID: 31018602 Free PMC article. Review.
-
Artificial Small RNA-Based Silencing Tools for Antiviral Resistance in Plants.Plants (Basel). 2020 May 26;9(6):669. doi: 10.3390/plants9060669. Plants (Basel). 2020. PMID: 32466363 Free PMC article. Review.
-
Mycovirus-encoded suppressors of RNA silencing: Possible allies or enemies in the use of RNAi to control fungal disease in crops.Front Fungal Biol. 2022 Oct 10;3:965781. doi: 10.3389/ffunb.2022.965781. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746227 Free PMC article. Review.
Cited by
-
Research Progress on miRNAs and Artificial miRNAs in Insect and Disease Resistance and Breeding in Plants.Genes (Basel). 2024 Sep 12;15(9):1200. doi: 10.3390/genes15091200. Genes (Basel). 2024. PMID: 39336791 Free PMC article. Review.
-
Plant biomarkers as early detection tools in stress management in food crops: a review.Planta. 2024 Feb 5;259(3):60. doi: 10.1007/s00425-024-04333-1. Planta. 2024. PMID: 38311674 Free PMC article. Review.
-
Rice microRNA156/529-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7/14/17 modules regulate defenses against bacteria.Plant Physiol. 2023 Jul 3;192(3):2537-2553. doi: 10.1093/plphys/kiad201. Plant Physiol. 2023. PMID: 36994827 Free PMC article.
-
Non-coding RNAs fine-tune the balance between plant growth and abiotic stress tolerance.Front Plant Sci. 2022 Oct 12;13:965745. doi: 10.3389/fpls.2022.965745. eCollection 2022. Front Plant Sci. 2022. PMID: 36311129 Free PMC article. Review.
-
Combined Omics Approaches Reveal Distinct Mechanisms of Resistance and/or Susceptibility in Sugar Beet Double Haploid Genotypes at Early Stages of Beet Curly Top Virus Infection.Int J Mol Sci. 2023 Oct 9;24(19):15013. doi: 10.3390/ijms241915013. Int J Mol Sci. 2023. PMID: 37834460 Free PMC article.
References
-
- Bao D, Ganbaatar O, Cui X, Yu R, Bao W, Falk BW, Wuriyanghan H. Down-regulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful soybean mosaic virus infection in soybean. Mol Plant Pathol. 2018;19:948–960. doi: 10.1111/mpp.12581. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

