SARS-CoV-2 prevalence in domestic and wildlife animals: A genomic and docking based structural comprehensive review
- PMID: 37662720
- PMCID: PMC10474441
- DOI: 10.1016/j.heliyon.2023.e19345
SARS-CoV-2 prevalence in domestic and wildlife animals: A genomic and docking based structural comprehensive review
Abstract
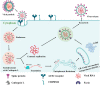
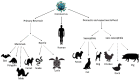
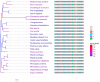
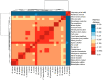
The SARS-CoV-2 virus has been identified as the infectious agent that led to the COVID-19 pandemic, which the world has seen very recently. Researchers have linked the SARS-CoV-2 outbreak to bats for the zoonotic spread of the virus to humans. Coronaviruses have a crown-like shape and positive-sense RNA nucleic acid. It attaches its spike glycoprotein to the host angiotensin-converting enzyme 2 (ACE2) receptor. Coronavirus genome comprises 14 ORFs and 27 proteins, spike glycoprotein being one of the most critical proteins for viral pathogenesis. Many mammals and reptiles, including bats, pangolins, ferrets, snakes, and turtles, serve as the principal reservoirs for this virus. But many experimental investigations have shown that certain domestic animals, including pigs, chickens, dogs, cats, and others, may also be able to harbor this virus, whether they exhibit any symptoms. These animals act as reservoirs for SARS-CoV, facilitating its zoonotic cross-species transmission to other species, including humans. In this review, we performed a phylogenetic analysis with multiple sequence alignment and pairwise evolutionary distance analysis, which revealed the similarity of ACE2 receptors in humans, chimpanzees, domestic rabbits, house mice, and golden hamsters. Pairwise RMSD analysis of the spike protein from some commonly reported SARS-CoV revealed that bat and pangolin coronavirus shared the highest structural similarity with human coronavirus. In a further experiment, molecular docking confirmed a higher affinity of pig, bat, and pangolin coronavirus spike proteins' affinity to the human ACE2 receptor. Such comprehensive structural and genomic analysis can help us to forecast the next likely animal source of these coronaviruses that may infect humans. To combat these zoonotic illnesses, we need a one health strategy that considers the well-being of people and animals and the local ecosystem.
Keywords: Mammals and reptiles; Molecular docking; One health strategy; Phylogenetic analysis; RMSD analysis; SARS-CoV-2.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


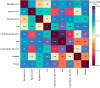

Similar articles
-
Molecular evolution and phylogenetic analysis of SARS-CoV-2 and hosts ACE2 protein suggest Malayan pangolin as intermediary host.Braz J Microbiol. 2020 Dec;51(4):1593-1599. doi: 10.1007/s42770-020-00321-1. Epub 2020 Jun 26. Braz J Microbiol. 2020. PMID: 32592038 Free PMC article.
-
SARS-CoV-2 and Three Related Coronaviruses Utilize Multiple ACE2 Orthologs and Are Potently Blocked by an Improved ACE2-Ig.J Virol. 2020 Oct 27;94(22):e01283-20. doi: 10.1128/JVI.01283-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847856 Free PMC article.
-
A SARS-CoV-2-Related Virus from Malayan Pangolin Causes Lung Infection without Severe Disease in Human ACE2-Transgenic Mice.J Virol. 2023 Feb 28;97(2):e0171922. doi: 10.1128/jvi.01719-22. Epub 2023 Jan 23. J Virol. 2023. PMID: 36688655 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19.Cells. 2021 Apr 6;10(4):821. doi: 10.3390/cells10040821. Cells. 2021. PMID: 33917481 Free PMC article. Review.
Cited by
-
Co-Circulation of Multiple Coronavirus Genera and Subgenera during an Epizootic of Lethal Respiratory Disease in Newborn Alpacas (Vicugna pacos) in Peru: First Report of Bat-like Coronaviruses in Alpacas.Animals (Basel). 2023 Sep 21;13(18):2983. doi: 10.3390/ani13182983. Animals (Basel). 2023. PMID: 37760383 Free PMC article.
-
Synthesis and in silico inhibitory action studies of azo-anchored imidazo[4,5-b]indole scaffolds against the COVID-19 main protease (Mpro).Sci Rep. 2024 May 6;14(1):10419. doi: 10.1038/s41598-024-57795-4. Sci Rep. 2024. PMID: 38710746 Free PMC article.
References
-
- Johansen M.D., Irving A., Montagutelli X., Tate M.D., Rudloff I., Nold M.F., Hansbro N.G., Kim R.Y., Donovan C., Liu G., Faiz A., Short K.R., Lyons J.G., McCaughan G.W., Gorrell M.D., Cole A., Moreno C., Couteur D., Hesselson D., Triccas J., Neely G.G., Gamble J.R., Simpson S.J., Saunders B.M., Oliver B.G., Britton W.J., Wark P.A., Nold-Petry C.A., Hansbro P.M. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020;13:877–891. doi: 10.1038/s41385-020-00340-z. - DOI - PMC - PubMed
-
- A.A. Rabaan, S.H. Al-Ahmed, S. Haque, R. Sah, R. Tiwari, Y.S. Malik, K. Dhama, M.I. Yatoo, D.K. Bonilla-Aldana, A.J. Rodriguez-Morales, SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview, Infez Med. 28 (n.d.) 174–184.. - PubMed
-
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.P., Pfenning A.R., Zhao H., Genereux D.P., Swofford R., Pollard K.S., Ryder O.A., Nweeia M.T., Lindblad-Toh K., Teeling E.C., Karlsson E.K., Lewin H.A. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

