Characterizing the transmembrane domains of ADAM10 and BACE1 and the impact of membrane composition
- PMID: 37658602
- PMCID: PMC10560698
- DOI: 10.1016/j.bpj.2023.08.025
Characterizing the transmembrane domains of ADAM10 and BACE1 and the impact of membrane composition
Abstract
The β-secretase, BACE1, and the α-secretase, ADAM10, are known to competitively cleave amyloid precursor protein (APP) in the amyloid cascades of Alzheimer's disease. Cleavage of APP by BACE1 produces a 99-residue C-terminal peptide (APP-C99) that is subsequently cleaved by γ-secretase to form amyloid-β (Aβ) protein, whereas cleavage of APP by ADAM10 is nonamyloidogenic. It has been speculated that ADAM10/APP and BACE1/APP interactions are regulated by colocalization within and outside of liquid-ordered membrane domains; however, the mechanism of this regulation and the character of the proteins' transmembrane domains are not well understood. In this work, we have developed and characterized minimal congener sequences for the transmembrane domains of ADAM10 and BACE1 using a multiscale modeling approach combining both temperature replica exchange and conventional molecular dynamics simulations based on the coarse-grained Martini2.2 and all-atom CHARMM36 force fields. Our results show that membrane composition impacts the character of the transmembrane domains of BACE1 and ADAM10, adding credence to the speculation that membrane domains are involved in the etiology of Alzheimer's disease.
Copyright © 2023 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




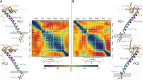
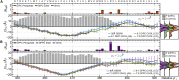
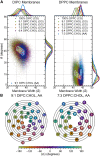
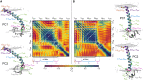
Similar articles
-
Proximity ligation assay reveals both pre- and postsynaptic localization of the APP-processing enzymes ADAM10 and BACE1 in rat and human adult brain.BMC Neurosci. 2020 Feb 4;21(1):6. doi: 10.1186/s12868-020-0554-0. BMC Neurosci. 2020. PMID: 32019490 Free PMC article.
-
Determination of the proteolytic cleavage sites of the amyloid precursor-like protein 2 by the proteases ADAM10, BACE1 and γ-secretase.PLoS One. 2011;6(6):e21337. doi: 10.1371/journal.pone.0021337. Epub 2011 Jun 17. PLoS One. 2011. PMID: 21695060 Free PMC article.
-
Alternative Selection of β-Site APP-Cleaving Enzyme 1 (BACE1) Cleavage Sites in Amyloid β-Protein Precursor (APP) Harboring Protective and Pathogenic Mutations within the Aβ Sequence.J Biol Chem. 2016 Nov 11;291(46):24041-24053. doi: 10.1074/jbc.M116.744722. Epub 2016 Sep 29. J Biol Chem. 2016. PMID: 27687728 Free PMC article.
-
Secretases in Alzheimer's disease: Novel insights into proteolysis of APP and TREM2.Curr Opin Neurobiol. 2022 Feb;72:101-110. doi: 10.1016/j.conb.2021.09.003. Epub 2021 Oct 22. Curr Opin Neurobiol. 2022. PMID: 34689040 Review.
-
BACE1: the beta-secretase enzyme in Alzheimer's disease.J Mol Neurosci. 2004;23(1-2):105-14. doi: 10.1385/JMN:23:1-2:105. J Mol Neurosci. 2004. PMID: 15126696 Review.
Cited by
-
Cholesterol and Lipid Rafts in the Biogenesis of Amyloid-β Protein and Alzheimer's Disease.Annu Rev Biophys. 2024 Jul;53(1):455-486. doi: 10.1146/annurev-biophys-062823-023436. Epub 2024 Jun 28. Annu Rev Biophys. 2024. PMID: 38382114 Review.
References
-
- Tolar M., Abushakra S., Sabbagh M. The path forward in Alzheimer’s disease therapeutics: Reevaluating the amyloid cascade hypothesis. Alzheimers Dement. 2020;16:1553–1560. - PubMed
-
- Baoukina S., Mendez-Villuendas E., et al. Tieleman D.P. Computer simulations of the phase separation in model membranes. Faraday Discuss. 2013;161:63–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

