Delayed gut microbiota maturation in the first year of life is a hallmark of pediatric allergic disease
- PMID: 37644001
- PMCID: PMC10465508
- DOI: 10.1038/s41467-023-40336-4
Delayed gut microbiota maturation in the first year of life is a hallmark of pediatric allergic disease
Abstract
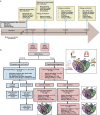
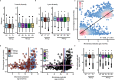
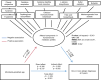
Allergic diseases affect millions of people worldwide. An increase in their prevalence has been associated with alterations in the gut microbiome, i.e., the microorganisms and their genes within the gastrointestinal tract. Maturation of the infant immune system and gut microbiota occur in parallel; thus, the conformation of the microbiome may determine if tolerant immune programming arises within the infant. Here we show, using deeply phenotyped participants in the CHILD birth cohort (n = 1115), that there are early-life influences and microbiome features which are uniformly associated with four distinct allergic diagnoses at 5 years: atopic dermatitis (AD, n = 367), asthma (As, n = 165), food allergy (FA, n = 136), and allergic rhinitis (AR, n = 187). In a subset with shotgun metagenomic and metabolomic profiling (n = 589), we discover that impaired 1-year microbiota maturation may be universal to pediatric allergies (AD p = 0.000014; As p = 0.0073; FA p = 0.00083; and AR p = 0.0021). Extending this, we find a core set of functional and metabolic imbalances characterized by compromised mucous integrity, elevated oxidative activity, decreased secondary fermentation, and elevated trace amines, to be a significant mediator between microbiota maturation at age 1 year and allergic diagnoses at age 5 years (βindirect = -2.28; p = 0.0020). Microbiota maturation thus provides a focal point to identify deviations from normative development to predict and prevent allergic disease.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Types and Amounts of Complementary Foods and Beverages and Food Allergy, Atopic Dermatitis/Eczema, Asthma, and Allergic Rhinitis: A Systematic Review [Internet].Alexandria (VA): USDA Nutrition Evidence Systematic Review; 2019 Apr. Alexandria (VA): USDA Nutrition Evidence Systematic Review; 2019 Apr. PMID: 35816599 Free Books & Documents. Review.
-
Relationship between Gut Microbiota and Allergies in Children: A Literature Review.Nutrients. 2023 May 29;15(11):2529. doi: 10.3390/nu15112529. Nutrients. 2023. PMID: 37299492 Free PMC article. Review.
-
Delayed Gut Colonization Shapes Future Allergic Responses in a Murine Model of Atopic Dermatitis.Front Immunol. 2021 Mar 17;12:650621. doi: 10.3389/fimmu.2021.650621. eCollection 2021. Front Immunol. 2021. PMID: 33815411 Free PMC article.
-
A Combined Analysis of Gut and Skin Microbiota in Infants with Food Allergy and Atopic Dermatitis: A Pilot Study.Nutrients. 2021 May 15;13(5):1682. doi: 10.3390/nu13051682. Nutrients. 2021. PMID: 34063398 Free PMC article.
-
Early life environmental exposure in relation to new onset and remission of allergic diseases in school children: Polish Mother and Child Cohort Study.Allergy Asthma Proc. 2019 Sep 1;40(5):329-337. doi: 10.2500/aap.2019.40.4237. Allergy Asthma Proc. 2019. PMID: 31514791
Cited by
-
Dietary Polyphenols-Natural Bioactive Compounds with Potential for Preventing and Treating Some Allergic Conditions.Nutrients. 2023 Nov 17;15(22):4823. doi: 10.3390/nu15224823. Nutrients. 2023. PMID: 38004216 Free PMC article. Review.
-
The Role of the Microbiota in the Pathogenesis and Treatment of Atopic Dermatitis-A Literature Review.Int J Mol Sci. 2024 Jun 13;25(12):6539. doi: 10.3390/ijms25126539. Int J Mol Sci. 2024. PMID: 38928245 Free PMC article. Review.
-
From womb to world: mapping gut microbiota-related health literacy among Italian mothers, a cross-sectional study.BMC Public Health. 2024 Apr 12;24(1):1012. doi: 10.1186/s12889-024-18497-8. BMC Public Health. 2024. PMID: 38605379 Free PMC article.
-
Experimental models of antibiotic exposure and atopic disease.Front Allergy. 2024 Oct 25;5:1455438. doi: 10.3389/falgy.2024.1455438. eCollection 2024. Front Allergy. 2024. PMID: 39525399 Free PMC article. Review.
-
Characterization of the gut microbiota and fecal and blood metabolomes under various factors in urban children from Northwest China.Front Cell Infect Microbiol. 2024 Mar 22;14:1374544. doi: 10.3389/fcimb.2024.1374544. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38585649 Free PMC article.
References
-
- Asher, M. I., García-Marcos, L., Pearce, N. E. & Strachan, D. P. Trends in worldwide asthma prevalence. Eur. Respir. J. 10.1183/13993003.02094-2020 (2020). - PubMed
-
- Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann. Nutr. Metab. 2015;66:8–16. - PubMed
-
- Petersen C, Turvey SE. Can we prevent allergic disease? Understanding the links between the early life microbiome and allergic diseases of childhood. Curr. Opin. Pediatr. 2020;32:790–797. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

