Epigenetic Regulation in Lean Nonalcoholic Fatty Liver Disease
- PMID: 37629043
- PMCID: PMC10454848
- DOI: 10.3390/ijms241612864
Epigenetic Regulation in Lean Nonalcoholic Fatty Liver Disease
Abstract
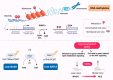
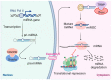
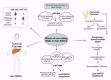
Nonalcoholic fatty liver disease (NAFLD), the most prominent cause of chronic liver disease worldwide, is a rapidly growing epidemic. It consists of a wide range of liver diseases, from steatosis to nonalcoholic steatohepatitis, and predisposes patients to liver fibrosis, cirrhosis, and even hepatocellular carcinoma. NAFLD is strongly correlated with obesity; however, it has been extensively reported among lean/nonobese individuals in recent years. Although lean patients demonstrate a lower prevalence of diabetes mellitus, central obesity, dyslipidemia, hypertension, and metabolic syndrome, a percentage of these patients may develop steatohepatitis, advanced liver fibrosis, and cardiovascular disease, and have increased all-cause mortality. The pathophysiological mechanisms of lean NAFLD remain vague. Studies have reported that lean NAFLD demonstrates a close association with environmental factors, genetic predisposition, and epigenetic modifications. In this review, we aim to discuss and summarize the epigenetic mechanisms involved in lean NAFLD and to introduce the interaction between epigenetic patterns and genetic or non genetic factors. Several epigenetic mechanisms have been implicated in the regulation of lean NAFLD. These include DNA methylation, histone modifications, and noncoding-RNA-mediated gene regulation. Epigenetics is an area of special interest in the setting of lean NAFLD as it could provide new insights into the therapeutic options and noninvasive biomarkers that target this under-recognized and challenging disorder.
Keywords: DNA methylation; epigenetic regulation; epigenetics; histone modification; lean NAFLD; nonalcoholic fatty liver disease; noncoding RNAs; nonobese NAFLD.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
AGA Clinical Practice Update: Diagnosis and Management of Nonalcoholic Fatty Liver Disease in Lean Individuals: Expert Review.Gastroenterology. 2022 Sep;163(3):764-774.e1. doi: 10.1053/j.gastro.2022.06.023. Epub 2022 Jul 14. Gastroenterology. 2022. PMID: 35842345 Free PMC article.
-
Recent advances in lean NAFLD.Biomed Pharmacother. 2022 Sep;153:113331. doi: 10.1016/j.biopha.2022.113331. Epub 2022 Jun 29. Biomed Pharmacother. 2022. PMID: 35779422 Review.
-
Histone Modifications in NAFLD: Mechanisms and Potential Therapy.Int J Mol Sci. 2023 Sep 27;24(19):14653. doi: 10.3390/ijms241914653. Int J Mol Sci. 2023. PMID: 37834101 Free PMC article. Review.
-
Special Population: Lean Nonalcoholic Fatty Liver Disease.Clin Liver Dis. 2023 May;27(2):451-469. doi: 10.1016/j.cld.2023.01.011. Epub 2023 Feb 26. Clin Liver Dis. 2023. PMID: 37024218 Review.
-
Noncoding RNAs as additional mediators of epigenetic regulation in nonalcoholic fatty liver disease.World J Gastroenterol. 2022 Sep 21;28(35):5111-5128. doi: 10.3748/wjg.v28.i35.5111. World J Gastroenterol. 2022. PMID: 36188722 Free PMC article. Review.
Cited by
-
Association between neutrophil-to-high-density lipoprotein cholesterol ratio and metabolic dysfunction-associated steatotic liver disease and liver fibrosis in the US population: a nationally representative cross-sectional study using NHANES data from 2017 to 2020.BMC Gastroenterol. 2024 Sep 5;24(1):300. doi: 10.1186/s12876-024-03394-6. BMC Gastroenterol. 2024. PMID: 39237899 Free PMC article.
-
Crosstalk between Epigenetics and Metabolic Reprogramming in Metabolic Dysfunction-Associated Steatotic Liver Disease-Induced Hepatocellular Carcinoma: A New Sight.Metabolites. 2024 Jun 8;14(6):325. doi: 10.3390/metabo14060325. Metabolites. 2024. PMID: 38921460 Free PMC article. Review.
-
Methylation status of LDLR, PCSK9 and LDLRAP1 is associated with cardiovascular events in familial hypercholesterolemia.Epigenomics. 2024;16(11-12):809-820. doi: 10.1080/17501911.2024.2351792. Epub 2024 Jun 17. Epigenomics. 2024. PMID: 38884343
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

