TP53 gain-of-function mutation modulates the immunosuppressive microenvironment in non-HPV-associated oral squamous cell carcinoma
- PMID: 37604640
- PMCID: PMC10445354
- DOI: 10.1136/jitc-2023-006666
TP53 gain-of-function mutation modulates the immunosuppressive microenvironment in non-HPV-associated oral squamous cell carcinoma
Abstract
Background: TP53, the most mutated gene in solid cancers, has a profound impact on most hallmarks of cancer. Somatic TP53 mutations occur in high frequencies in head and neck cancers, including oral squamous cell carcinoma (OSCC). Our study aims to understand the role of TP53 gain-of-function mutation in modulating the tumor immune microenvironment (TIME) in OSCC.
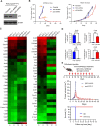
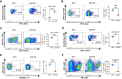
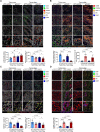
Methods: Short hairpin RNA knockdown of mutant p53R172H in syngeneic oral tumors demonstrated changes in tumor growth between immunocompetent and immunodeficient mice. HTG EdgeSeq targeted messenger RNA sequencing was used to analyze cytokine and immune cell markers in tumors with inactivated mutant p53R172H. Flow cytometry and multiplex immunofluorescence (mIF) confirmed the role of mutant p53R172H in the TIME. The gene expression of patients with OSCC was analyzed by CIBERSORT and mIF was used to validate the immune landscape at the protein level.
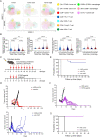
Results: Mutant p53R172H contributes to a cytokine transcriptome network that inhibits the infiltration of cytotoxic CD8+ T cells and promotes intratumoral recruitment of regulatory T cells and M2 macrophages. Moreover, p53R172H also regulates the spatial distribution of immunocyte populations, and their distribution between central and peripheral intratumoral locations. Interestingly, p53R172H-mutated tumors are infiltrated with CD8+ and CD4+ T cells expressing programmed cell death protein 1, and these tumors responded to immune checkpoint inhibitor and stimulator of interferon gene 1 agonist therapy. CIBERSORT analysis of human OSCC samples revealed associations between immune cell populations and the TP53R175H mutation, which paralleled the findings from our syngeneic mouse tumor model.
Conclusions: These findings demonstrate that syngeneic tumors bearing the TP53R172H gain-of-function mutation modulate the TIME to evade tumor immunity, leading to tumor progression and decreased survival.
Keywords: cytokines; head and neck neoplasms; immune checkpoint inhibitors; immunotherapy; tumor microenvironment.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures

Similar articles
-
Mutant p53 drives an immune cold tumor immune microenvironment in oral squamous cell carcinoma.Commun Biol. 2022 Jul 28;5(1):757. doi: 10.1038/s42003-022-03675-4. Commun Biol. 2022. PMID: 35902768 Free PMC article.
-
The immune microenvironment of HPV-positive and HPV-negative oropharyngeal squamous cell carcinoma: a multiparametric quantitative and spatial analysis unveils a rationale to target treatment-naïve tumors with immune checkpoint inhibitors.J Exp Clin Cancer Res. 2022 Sep 20;41(1):279. doi: 10.1186/s13046-022-02481-4. J Exp Clin Cancer Res. 2022. PMID: 36123711 Free PMC article.
-
HPV-positive murine oral squamous cell carcinoma: development and characterization of a new mouse tumor model for immunological studies.J Transl Med. 2023 Jun 10;21(1):376. doi: 10.1186/s12967-023-04221-4. J Transl Med. 2023. PMID: 37296466 Free PMC article.
-
Expression of SESN1, UHRF1BP1, and miR-377-3p as prognostic markers in mutated TP53 squamous cell carcinoma of the head and neck.Cancer Biol Ther. 2017 Oct 3;18(10):775-782. doi: 10.1080/15384047.2017.1373212. Epub 2017 Sep 8. Cancer Biol Ther. 2017. PMID: 28886272 Free PMC article. Review.
-
Tumor-infiltrating ICOS+ Effector Regulatory T-Cells in Oral Squamous Cell Carcinoma as a Promising Biomarker for Prognosis and 'Hot' Tumor.Anticancer Res. 2022 May;42(5):2383-2393. doi: 10.21873/anticanres.15717. Anticancer Res. 2022. PMID: 35489733 Review.
Cited by
-
Machine learning developed a macrophage signature for predicting prognosis, immune infiltration and immunotherapy features in head and neck squamous cell carcinoma.Sci Rep. 2024 Aug 22;14(1):19538. doi: 10.1038/s41598-024-70430-6. Sci Rep. 2024. PMID: 39174693 Free PMC article.
References
-
- Califano J, van der Riet P, Westra W, et al. . Genetic progression model for head and neck cancer: implications for field Cancerization. Cancer Res 1996;56:2488–92. - PubMed
-
- Califano J, Westra WH, Meininger G, et al. . Genetic progression and Clonal relationship of recurrent Premalignant head and neck lesions. Clin Cancer Res 2000;6:347–52. - PubMed
-
- Myers J. Cancer of the head and neck. 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
