On the micelle formation of DNAJB6b
- PMID: 37593255
- PMCID: PMC10427797
- DOI: 10.1017/qrd.2023.4
On the micelle formation of DNAJB6b
Abstract
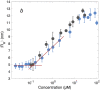
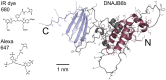
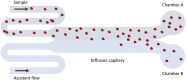
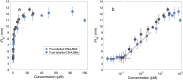
The human chaperone DNAJB6b increases the solubility of proteins involved in protein aggregation diseases and suppresses the nucleation of amyloid structures. Due to such favourable properties, DNAJB6b has gained increasing attention over the past decade. The understanding of how DNAJB6b operates on a molecular level may aid the design of inhibitors against amyloid formation. In this work, fundamental aspects of DNAJB6b self-assembly have been examined, providing a basis for future experimental designs and conclusions. The results imply the formation of large chaperone clusters in a concentration-dependent manner. Microfluidic diffusional sizing (MDS) was used to evaluate how DNAJB6b average hydrodynamic radius varies with concentration. We found that, in 20 mM sodium phosphate buffer, 0.2 mM EDTA, at pH 8.0 and room temperature, DNAJB6b displays a micellar behaviour, with a critical micelle concentration (CMC) of around 120 nM. The average hydrodynamic radius appears to be concentration independent between ∼10 μM and 100 μM, with a mean radius of about 12 nm. The CMC found by MDS is supported by native agarose gel electrophoresis and the size distribution appears bimodal in the DNAJB6b concentration range ∼100 nM to 4 μM.
Keywords: affinity; aggregation; chaperone action; oligomers; self-association.
© The Author(s) 2023.
Figures


Similar articles
-
The Ability of DNAJB6b to Suppress Amyloid Formation Depends on the Chaperone Aggregation State.ACS Chem Neurosci. 2024 May 1;15(9):1732-1737. doi: 10.1021/acschemneuro.4c00120. Epub 2024 Apr 19. ACS Chem Neurosci. 2024. PMID: 38640082 Free PMC article.
-
High-Efficiency Expression and Purification of DNAJB6b Based on the pH-Modulation of Solubility and Denaturant-Modulation of Size.Molecules. 2022 Jan 10;27(2):418. doi: 10.3390/molecules27020418. Molecules. 2022. PMID: 35056736 Free PMC article.
-
The HSP40 family chaperone isoform DNAJB6b prevents neuronal cells from tau aggregation.BMC Biol. 2023 Dec 18;21(1):293. doi: 10.1186/s12915-023-01798-6. BMC Biol. 2023. PMID: 38110916 Free PMC article.
-
Gel filtration chromatographic study on the self-association of surfactants and related compounds.Adv Colloid Interface Sci. 1993 Jan 25;43(1):87-136. doi: 10.1016/0001-8686(93)80006-w. Adv Colloid Interface Sci. 1993. PMID: 7680872 Review.
-
Proteostasis and the Regulation of Intra- and Extracellular Protein Aggregation by ATP-Independent Molecular Chaperones: Lens α-Crystallins and Milk Caseins.Acc Chem Res. 2018 Mar 20;51(3):745-752. doi: 10.1021/acs.accounts.7b00250. Epub 2018 Feb 14. Acc Chem Res. 2018. PMID: 29442498 Review.
Cited by
-
The Ability of DNAJB6b to Suppress Amyloid Formation Depends on the Chaperone Aggregation State.ACS Chem Neurosci. 2024 May 1;15(9):1732-1737. doi: 10.1021/acschemneuro.4c00120. Epub 2024 Apr 19. ACS Chem Neurosci. 2024. PMID: 38640082 Free PMC article.
References
-
- Arkan S, Ljungberg M, Kirik D and Hansen C (2021) DNAJB6 suppresses alphasynuclein induced pathology in an animal model of Parkinson’s disease. Neurobiology of Disease 158, 105477. - PubMed
-
- Baldwin AJ, Lioe H, Robinson CV, Kay LE and Benesch JL (2011) αb-crystallin polydispersity is a consequence of unbiased quaternary dynamics. Journal of Molecular Biology 413(2), 297–309. - PubMed
-
- Cantor RC and Schimmel RP (1980) Biophysical Chemistry Part II:Techniques for the Study of Biological Structure and Function. San Francisco: W. H. Freeman and Company.
-
- Chien V, Aitken JF, Zhang S, Buchanan CM, Hickey A, Brittain T, Cooper GJ and Loomes KM (2010) The chaperone proteins HSP70, HSP40/DnaJ and GRP78/BiP suppress misfolding and formation of β-sheet-containing aggregates by human amylin: A potential role for defective chaperone biology in type 2 diabetes. Biochemical Journal 432(1), 113–121. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
