Reconstitution of phytochrome A-mediated light modulation of the ABA signaling pathways in yeast
- PMID: 37590408
- PMCID: PMC10450666
- DOI: 10.1073/pnas.2302901120
Reconstitution of phytochrome A-mediated light modulation of the ABA signaling pathways in yeast
Abstract
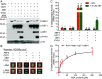
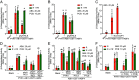
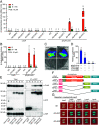
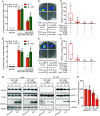
Abscisic acid (ABA), a classical plant hormone, plays an essential role in plant adaptation to environmental stresses. The ABA signaling mechanisms have been extensively investigated, and it was shown that the PYR1 (PYRABACTIN RESISTANCE1)/PYL (PYR1-LIKE)/RCAR (REGULATORY COMPONENT OF ABA RECEPTOR) ABA receptors, the PP2C coreceptors, and the SnRK2 protein kinases constitute the core ABA signaling module responsible for ABA perception and initiation of downstream responses. We recently showed that ABA signaling is modulated by light signals, but the underlying molecular mechanisms remain largely obscure. In this study, we established a system in yeast cells that was not only successful in reconstituting a complete ABA signaling pathway, from hormone perception to ABA-responsive gene expression, but also suitable for functionally characterizing the regulatory roles of additional factors of ABA signaling. Using this system, we analyzed the roles of several light signaling components, including the red and far-red light photoreceptors phytochrome A (phyA) and phyB, and the photomorphogenic central repressor COP1, in the regulation of ABA signaling. Our results showed that both phyA and phyB negatively regulated ABA signaling, whereas COP1 positively regulated ABA signaling in yeast cells. Further analyses showed that photoactivated phyA interacted with the ABA coreceptors ABI1 and ABI2 to decrease their interactions with the ABA receptor PYR1. Together, data from our reconstituted yeast ABA signaling system provide evidence that photoactivated photoreceptors attenuate ABA signaling by directly interacting with the key components of the core ABA signaling module, thus conferring enhanced ABA tolerance to light-grown plants.
Keywords: ABA; light; phyA; reconstitution; yeast.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action.Proc Natl Acad Sci U S A. 2012 Jun 26;109(26):10593-8. doi: 10.1073/pnas.1116590109. Epub 2012 Jun 11. Proc Natl Acad Sci U S A. 2012. PMID: 22689970 Free PMC article.
-
PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis.Plant J. 2010 Jan;61(2):290-9. doi: 10.1111/j.1365-313X.2009.04054.x. Epub 2009 Oct 26. Plant J. 2010. PMID: 19874541 Free PMC article.
-
Phytochrome A and B Function Antagonistically to Regulate Cold Tolerance via Abscisic Acid-Dependent Jasmonate Signaling.Plant Physiol. 2016 Jan;170(1):459-71. doi: 10.1104/pp.15.01171. Epub 2015 Nov 2. Plant Physiol. 2016. PMID: 26527654 Free PMC article.
-
Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants.Physiol Plant. 2013 Jan;147(1):15-27. doi: 10.1111/j.1399-3054.2012.01635.x. Epub 2012 May 16. Physiol Plant. 2013. PMID: 22519646 Review.
-
Evolution of Abscisic Acid Signaling Module and Its Perception.Front Plant Sci. 2020 Jul 10;11:934. doi: 10.3389/fpls.2020.00934. eCollection 2020. Front Plant Sci. 2020. PMID: 32754170 Free PMC article. Review.
Cited by
-
Osmotic signaling releases PP2C-mediated inhibition of Arabidopsis SnRK2s via the receptor-like cytoplasmic kinase BIK1.EMBO J. 2024 Oct 21. doi: 10.1038/s44318-024-00277-0. Online ahead of print. EMBO J. 2024. PMID: 39433899
-
Arabidopsis MYB21 Negatively Regulates KTN1 to Fine-Tune the Filament Elongation.Plants (Basel). 2023 Nov 17;12(22):3884. doi: 10.3390/plants12223884. Plants (Basel). 2023. PMID: 38005781 Free PMC article.
References
-
- Chen X., et al. , Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 63, 53–78 (2021). - PubMed
-
- Chen K., et al. , Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54 (2020). - PubMed
-
- Gong Z., et al. , Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674 (2020). - PubMed
-
- Zhang H., Zhu J., Gong Z., Zhu J. K., Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119 (2022). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

