Exploring New Dimensions of Tumor Heterogeneity: The Application of Single Cell Analysis to Organoid-Based 3D In Vitro Models
- PMID: 37589373
- PMCID: PMC11468421
- DOI: 10.1002/adhm.202300903
Exploring New Dimensions of Tumor Heterogeneity: The Application of Single Cell Analysis to Organoid-Based 3D In Vitro Models
Abstract
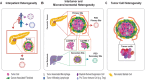
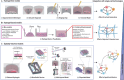
Modeling the heterogeneity of the tumor microenvironment (TME) in vitro is essential to investigating fundamental cancer biology and developing novel treatment strategies that holistically address the factors affecting tumor progression and therapeutic response. Thus, the development of new tools for both in vitro modeling, such as patient-derived organoids (PDOs) and complex 3D in vitro models, and single cell omics analysis, such as single-cell RNA-sequencing, represents a new frontier for investigating tumor heterogeneity. Specifically, the integration of PDO-based 3D in vitro models and single cell analysis offers a unique opportunity to explore the intersecting effects of interpatient, microenvironmental, and tumor cell heterogeneity on cell phenotypes in the TME. In this review, the current use of PDOs in complex 3D in vitro models of the TME is discussed and the emerging directions in the development of these models are highlighted. Next, work that has successfully applied single cell analysis to PDO-based models is examined and important experimental considerations are identified for this approach. Finally, open questions are highlighted that may be amenable to exploration using the integration of PDO-based models and single cell analysis. Ultimately, such investigations may facilitate the identification of novel therapeutic targets for cancer that address the significant influence of tumor-TME interactions.
Keywords: 3D in vitro models; single cell transcriptomics; tissue engineering; tumor heterogeneity; tumor microenvironments.
© 2023 The Authors. Advanced Healthcare Materials published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Patient derived organoids in prostate cancer: improving therapeutic efficacy in precision medicine.Mol Cancer. 2021 Sep 29;20(1):125. doi: 10.1186/s12943-021-01426-3. Mol Cancer. 2021. PMID: 34587953 Free PMC article. Review.
-
Tumor organoid models in precision medicine and investigating cancer-stromal interactions.Pharmacol Ther. 2021 Feb;218:107668. doi: 10.1016/j.pharmthera.2020.107668. Epub 2020 Aug 24. Pharmacol Ther. 2021. PMID: 32853629 Free PMC article. Review.
-
Patient-derived organoid culture in epithelial ovarian cancers-Techniques, applications, and future perspectives.Cancer Med. 2023 Oct;12(19):19714-19731. doi: 10.1002/cam4.6521. Epub 2023 Sep 30. Cancer Med. 2023. PMID: 37776168 Free PMC article. Review.
-
Organoid Modeling of the Tumor Immune Microenvironment.Cell. 2018 Dec 13;175(7):1972-1988.e16. doi: 10.1016/j.cell.2018.11.021. Cell. 2018. PMID: 30550791 Free PMC article.
-
Transcriptomic intratumor heterogeneity of breast cancer patient-derived organoids may reflect the unique biological features of the tumor of origin.Breast Cancer Res. 2023 Feb 21;25(1):21. doi: 10.1186/s13058-023-01617-4. Breast Cancer Res. 2023. PMID: 36810117 Free PMC article.
Cited by
-
Bio-orthogonal tuning of matrix properties during 3D cell culture to induce morphological and phenotypic changes.Nat Protoc. 2024 Nov 5. doi: 10.1038/s41596-024-01066-z. Online ahead of print. Nat Protoc. 2024. PMID: 39501109 Review.
-
Innovations in three-dimensional-printed individualized bone prosthesis materials: revolutionizing orthopedic surgery: a review.Int J Surg. 2024 Oct 1;110(10):6748-6762. doi: 10.1097/JS9.0000000000001842. Int J Surg. 2024. PMID: 38905508 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

