A low-background, fluorescent assay to evaluate inhibitors of diverse viral proteases
- PMID: 37578235
- PMCID: PMC10506478
- DOI: 10.1128/jvi.00597-23
A low-background, fluorescent assay to evaluate inhibitors of diverse viral proteases
Abstract
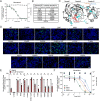
Multiple coronaviruses (CoVs) can cause respiratory diseases in humans. While prophylactic vaccines designed to prevent infection are available for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), incomplete vaccine efficacy, vaccine hesitancy, and the threat of other pathogenic CoVs for which vaccines do not exist have highlighted the need for effective antiviral therapies. While antiviral compounds targeting the viral polymerase and protease are already in clinical use, their sensitivity to potential resistance mutations as well as their breadth against the full range of human and preemergent CoVs remain incompletely defined. To begin to fill that gap in knowledge, we report here the development of an improved, noninfectious, cell-based fluorescent assay with high sensitivity and low background that reports on the activity of viral proteases, which are key drug targets. We demonstrate that the assay is compatible with not only the SARS-CoV-2 Mpro protein but also orthologues from a range of human and nonhuman CoVs as well as clinically reported SARS-CoV-2 drug-resistant Mpro variants. We then use this assay to define the breadth of activity of two clinically used protease inhibitors, nirmatrelvir and ensitrelvir. Continued use of this assay will help define the strengths and limitations of current therapies and may also facilitate the development of next-generation protease inhibitors that are broadly active against both currently circulating and preemergent CoVs. IMPORTANCE Coronaviruses (CoVs) are important human pathogens with the ability to cause global pandemics. Working in concert with vaccines, antivirals specifically limit viral disease in people who are actively infected. Antiviral compounds that target CoV proteases are already in clinical use; their efficacy against variant proteases and preemergent zoonotic CoVs, however, remains incompletely defined. Here, we report an improved, noninfectious, and highly sensitive fluorescent method of defining the sensitivity of CoV proteases to small molecule inhibitors. We use this approach to assay the activity of current antiviral therapies against clinically reported SARS-CoV-2 protease mutants and a panel of highly diverse CoV proteases. Additionally, we show this system is adaptable to other structurally nonrelated viral proteases. In the future, this assay can be used to not only better define the strengths and limitations of current therapies but also help develop new, broadly acting inhibitors that more broadly target viral families.
Keywords: CoV Mpro; CoV PLpro; Epstein–Barr virus; FlipGFP; Zika virus; antivirals; coronaviruses.
Conflict of interest statement
Some authors have IP on viral protease reporter technology.
Figures



Similar articles
-
Development of a Fluorescence-Based, High-Throughput SARS-CoV-2 3CLpro Reporter Assay.J Virol. 2020 Oct 27;94(22):e01265-20. doi: 10.1128/JVI.01265-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32843534 Free PMC article.
-
Adaptive Mutation in the Main Protease Cleavage Site of Feline Coronavirus Renders the Virus More Resistant to Main Protease Inhibitors.J Virol. 2022 Sep 14;96(17):e0090722. doi: 10.1128/jvi.00907-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000844 Free PMC article.
-
Genetic Surveillance of SARS-CoV-2 Mpro Reveals High Sequence and Structural Conservation Prior to the Introduction of Protease Inhibitor Paxlovid.mBio. 2022 Aug 30;13(4):e0086922. doi: 10.1128/mbio.00869-22. Epub 2022 Jul 13. mBio. 2022. PMID: 35862764 Free PMC article.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy.ACS Comb Sci. 2020 Jun 8;22(6):297-305. doi: 10.1021/acscombsci.0c00058. Epub 2020 May 27. ACS Comb Sci. 2020. PMID: 32402186 Review.
References
-
- Iketani S, Liu L, Guo Y, Liu L, Chan J-W, Huang Y, Wang M, Luo Y, Yu J, Chu H, Chik K-H, Yuen T-T, Yin MT, Sobieszczyk ME, Huang Y, Yuen K-Y, Wang HH, Sheng Z, Ho DD. 2022. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604:553–556. doi:10.1038/s41586-022-04594-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

