Preliminary landscape of Candidatus Saccharibacteria in the human microbiome
- PMID: 37577371
- PMCID: PMC10414567
- DOI: 10.3389/fcimb.2023.1195679
Preliminary landscape of Candidatus Saccharibacteria in the human microbiome
Abstract
Introduction: Candidate Phyla Radiation (CPR) and more specifically Candidatus Saccharibacteria (TM7) have now been established as ubiquitous members of the human oral microbiota. Additionally, CPR have been reported in the gastrointestinal and urogenital tracts. However, the exploration of new human niches has been limited to date.
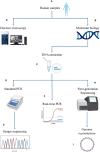
Methods: In this study, we performed a prospective and retrospective screening of TM7 in human samples using standard PCR, real-time PCR, scanning electron microscopy (SEM) and shotgun metagenomics.
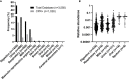
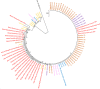
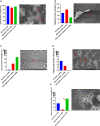
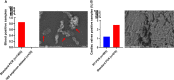
Results: Using Real-time PCR and standard PCR, oral samples presented the highest TM7 prevalence followed by fecal samples, breast milk samples, vaginal samples and urine samples. Surprisingly, TM7 were also detected in infectious samples, namely cardiac valves and blood cultures at a low prevalence (under 3%). Moreover, we observed CPR-like structures using SEM in all sample types except cardiac valves. The reconstruction of TM7 genomes in oral and fecal samples from shotgun metagenomics reads further confirmed their high prevalence in some samples.
Conclusion: This study confirmed, through their detection in multiple human samples, that TM7 are human commensals that can also be found in clinical settings. Their detection in clinical samples warrants further studies to explore their role in a pathological setting.
Keywords: Candidate Phyla Radiation; Candidatus Saccharibacteria; electron microscopy; human microbiome; molecular detection.
Copyright © 2023 Naud, Valles, Abdillah, Abou Chacra, Mekhalif, Ibrahim, Caputo, Baudoin, Gouriet, Bittar, Lagier, Ranque, Fenollar, Tidjani Alou and Raoult.
Conflict of interest statement
DR was a consultant in microbiology for the Hitachi High-Tech Corporation until March 2021. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be misconstrued as a potential conflict of interest.
Figures
Similar articles
-
Adapted Protocol for Saccharibacteria Cocultivation: Two New Members Join the Club of Candidate Phyla Radiation.Microbiol Spectr. 2021 Dec 22;9(3):e0106921. doi: 10.1128/spectrum.01069-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 35007432 Free PMC article.
-
Strain-Level Variation and Diverse Host Bacterial Responses in Episymbiotic Saccharibacteria.mSystems. 2022 Apr 26;7(2):e0148821. doi: 10.1128/msystems.01488-21. Epub 2022 Mar 28. mSystems. 2022. PMID: 35343799 Free PMC article.
-
Structure and Composition of Candidate Phyla Radiation in Supragingival Plaque of Caries Patients.Chin J Dent Res. 2022 Jun 10;25(2):107-118. doi: 10.3290/j.cjdr.b3086339. Chin J Dent Res. 2022. PMID: 35686590
-
Saccharibacteria (TM7) in the Human Oral Microbiome.J Dent Res. 2019 May;98(5):500-509. doi: 10.1177/0022034519831671. Epub 2019 Mar 20. J Dent Res. 2019. PMID: 30894042 Free PMC article. Review.
-
Candidate Phyla Radiation, an Underappreciated Division of the Human Microbiome, and Its Impact on Health and Disease.Clin Microbiol Rev. 2022 Sep 21;35(3):e0014021. doi: 10.1128/cmr.00140-21. Epub 2022 Jun 6. Clin Microbiol Rev. 2022. PMID: 35658516 Free PMC article. Review.
Cited by
-
From wolves to humans: oral microbiome resistance to transfer across mammalian hosts.mBio. 2024 Mar 13;15(3):e0334223. doi: 10.1128/mbio.03342-23. Epub 2024 Feb 1. mBio. 2024. PMID: 38299854 Free PMC article.
-
Periodontitis: etiology, conventional treatments, and emerging bacteriophage and predatory bacteria therapies.Front Microbiol. 2024 Sep 26;15:1469414. doi: 10.3389/fmicb.2024.1469414. eCollection 2024. Front Microbiol. 2024. PMID: 39391608 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

