Amniotic Membrane-Derived Stromal Cells Release Extracellular Vesicles That Favor Regeneration of Dystrophic Skeletal Muscles
- PMID: 37569832
- PMCID: PMC10418925
- DOI: 10.3390/ijms241512457
Amniotic Membrane-Derived Stromal Cells Release Extracellular Vesicles That Favor Regeneration of Dystrophic Skeletal Muscles
Abstract
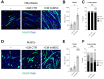
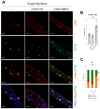
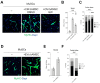
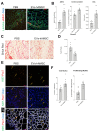
Duchenne muscular dystrophy (DMD) is a muscle disease caused by mutations in the dystrophin gene characterized by myofiber fragility and progressive muscle degeneration. The genetic defect results in a reduced number of self-renewing muscle stem cells (MuSCs) and an impairment of their activation and differentiation, which lead to the exhaustion of skeletal muscle regeneration potential and muscle replacement by fibrotic and fatty tissue. In this study, we focused on an unexplored strategy to improve MuSC function and to preserve their niche based on the regenerative properties of mesenchymal stromal cells from the amniotic membrane (hAMSCs), that are multipotent cells recognized to have a role in tissue repair in different disease models. We demonstrate that the hAMSC secretome (CM hAMSC) and extracellular vesicles (EVs) isolated thereof directly stimulate the in vitro proliferation and differentiation of human myoblasts and mouse MuSC from dystrophic muscles. Furthermore, we demonstrate that hAMSC secreted factors modulate the muscle stem cell niche in dystrophic-mdx-mice. Interestingly, local injection of EV hAMSC in mdx muscles correlated with an increase in the number of activated Pax7+/Ki67+ MuSCs and in new fiber formation. EV hAMSCs also significantly reduced muscle collagen deposition, thus counteracting fibrosis and MuSCs exhaustion, two hallmarks of DMD. Herein for the first time we demonstrate that CM hAMSC and EVs derived thereof promote muscle regeneration by supporting proliferation and differentiation of resident muscle stem cells. These results pave the way for the development of a novel treatment to counteract DMD progression by reducing fibrosis and enhancing myogenesis in dystrophic muscles.
Keywords: Duchenne muscular dystrophy; amnion; extracellular vesicles; mesenchymal stem/stromal cells; skeletal muscle regeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Immunomodulatory amnion-derived mesenchymal stromal cells preserve muscle function in a mouse model of Duchenne muscular dystrophy.Stem Cell Res Ther. 2023 Apr 27;14(1):108. doi: 10.1186/s13287-023-03337-0. Stem Cell Res Ther. 2023. PMID: 37106393 Free PMC article.
-
Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy.Exp Physiol. 2014 Apr;99(4):675-87. doi: 10.1113/expphysiol.2013.077255. Epub 2014 Jan 17. Exp Physiol. 2014. PMID: 24443351
-
MyD88 is required for satellite cell-mediated myofiber regeneration in dystrophin-deficient mdx mice.Hum Mol Genet. 2018 Oct 1;27(19):3449-3463. doi: 10.1093/hmg/ddy258. Hum Mol Genet. 2018. PMID: 30010933 Free PMC article.
-
Therapeutic Application of Extracellular Vesicles-Capsulated Adeno-Associated Virus Vector via nSMase2/Smpd3, Satellite, and Immune Cells in Duchenne Muscular Dystrophy.Int J Mol Sci. 2022 Jan 28;23(3):1551. doi: 10.3390/ijms23031551. Int J Mol Sci. 2022. PMID: 35163475 Free PMC article. Review.
-
Satellite Cells in Muscular Dystrophy - Lost in Polarity.Trends Mol Med. 2016 Jun;22(6):479-496. doi: 10.1016/j.molmed.2016.04.002. Epub 2016 May 5. Trends Mol Med. 2016. PMID: 27161598 Free PMC article. Review.
References
-
- Giampà C., Alvino A., Magatti M., Silini A.R., Cardinale A., Paldino E., Fusco F.R., Parolini O. Conditioned medium from amniotic cells protects striatal degeneration and ameliorates motor deficits in the R6/2 mouse model of Huntington’s disease. J. Cell Mol. Med. 2019;23:1581–1592. doi: 10.1111/jcmm.14113. - DOI - PMC - PubMed
-
- Ratajczak M.Z., Kucia M., Jadczyk T., Greco N.J., Wojakowski W., Tendera M., Ratajczak J. Pivotal role of paracrine effects in stem cell therapies in regenerative medicine: Can we translate stem cell-secreted paracrine factors and microvesicles into better therapeutic strategies? Leukemia. 2021;26:1166–1173. doi: 10.1038/leu.2011.389. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

