The Origin of Highly Elevated Cell-Free DNA in Healthy Individuals and Patients with Pancreatic, Colorectal, Lung, or Ovarian Cancer
- PMID: 37565753
- PMCID: PMC10592331
- DOI: 10.1158/2159-8290.CD-21-1252
The Origin of Highly Elevated Cell-Free DNA in Healthy Individuals and Patients with Pancreatic, Colorectal, Lung, or Ovarian Cancer
Abstract
Cell-free DNA (cfDNA) concentrations from patients with cancer are often elevated compared with those of healthy controls, but the sources of this extra cfDNA have never been determined. To address this issue, we assessed cfDNA methylation patterns in 178 patients with cancers of the colon, pancreas, lung, or ovary and 64 patients without cancer. Eighty-three of these individuals had cfDNA concentrations much greater than those generally observed in healthy subjects. The major contributor of cfDNA in all samples was leukocytes, accounting for ∼76% of cfDNA, with neutrophils predominating. This was true regardless of whether the samples were derived from patients with cancer or the total plasma cfDNA concentration. High levels of cfDNA observed in patients with cancer did not come from either neoplastic cells or surrounding normal epithelial cells from the tumor's tissue of origin. These data suggest that cancers may have a systemic effect on cell turnover or DNA clearance.
Significance: The origin of excess cfDNA in patients with cancer is unknown. Using cfDNA methylation patterns, we determined that neither the tumor nor the surrounding normal tissue contributes this excess cfDNA-rather it comes from leukocytes. This finding suggests that cancers have a systemic impact on cell turnover or DNA clearance. See related commentary by Thierry and Pisareva, p. 2122. This article is featured in Selected Articles from This Issue, p. 2109.
©2023 American Association for Cancer Research.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
BV, KWK, & NP are founders of Thrive Earlier Detection, an Exact Sciences Company. BV, KWK, NP, and CD hold equity in Exact Sciences. BV, KWK, and NP are founders of Personal Genome Diagnostics. KWK, BV, & NP hold equity in are consultants to CAGE Pharma. KWK, and NP own equity in Neophore and KWK and NP are consultants to Neophore. BV is a consultant to and holds equity in Catalio Capital Management. CB is a consultant to Depuy-Synthes and Bionaut Labs. The companies named above, as well as other companies, have licensed previously described technologies related to the work described in this paper from Johns Hopkins University. BV, KWK, NP, CB, RH, CT, CD, AKM, and JDC are inventors on some of these technologies. Licenses to these technologies are or will be associated with equity or royalty payments to the inventors as well as to Johns Hopkins University. Patent applications on the work described in this paper may be filed by Johns Hopkins University. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict-of-interest policies. YMDL is a scientific cofounder, past member of scientific advisory board and past consultant of Grail. YMDL and KCAC hold equities and are board members of Take2 and DRA Limited. YMDL, KCAC and PJ receive patent licensing incomes from Illumina, Sequenom, Xcelom, Grail, Take2 and DRA Limited. KCAC is a past consultant to Grail. PJ holds equities in Grail. PJ is a consultant of Take2 Technologies Limited. PJ is a Director of KingMed Future.
Figures
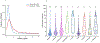
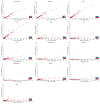


Comment in
-
A New Paradigm of the Origins of Circulating DNA in Patients with Cancer.Cancer Discov. 2023 Oct 5;13(10):2122-2124. doi: 10.1158/2159-8290.CD-23-0824. Cancer Discov. 2023. PMID: 37794839
References
-
- Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem 2002;48(3):421–7. - PubMed
-
- Mandel P, Metais P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil 1948;142(3-4):241–3. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

