Human cytomegalovirus mediates APOBEC3B relocalization early during infection through a ribonucleotide reductase-independent mechanism
- PMID: 37565748
- PMCID: PMC10506462
- DOI: 10.1128/jvi.00781-23
Human cytomegalovirus mediates APOBEC3B relocalization early during infection through a ribonucleotide reductase-independent mechanism
Abstract
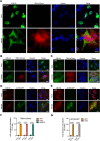
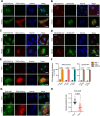
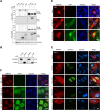
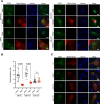
The APOBEC3 family of DNA cytosine deaminases comprises an important arm of the innate antiviral defense system. The gamma-herpesviruses Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus and the alpha-herpesviruses herpes simplex virus (HSV)-1 and HSV-2 have evolved an efficient mechanism to avoid APOBEC3 restriction by directly binding to APOBEC3B and facilitating its exclusion from the nuclear compartment. The only viral protein required for APOBEC3B relocalization is the large subunit of the ribonucleotide reductase (RNR). Here, we ask whether this APOBEC3B relocalization mechanism is conserved with the beta-herpesvirus human cytomegalovirus (HCMV). Although HCMV infection causes APOBEC3B relocalization from the nucleus to the cytoplasm in multiple cell types, the viral RNR (UL45) is not required. APOBEC3B relocalization occurs rapidly following infection suggesting the involvement of an immediate early or early (IE/E) viral protein. In support of this possibility, genetic (IE1 mutant) and pharmacologic (cycloheximide) strategies that prevent the expression of IE/E viral proteins also block APOBEC3B relocalization. In comparison, the treatment of infected cells with phosphonoacetic acid, which interferes with viral late protein expression, still permits A3B relocalization. These results combine to indicate that the beta-herpesvirus HCMV uses an RNR-independent, yet phenotypically similar, molecular mechanism to antagonize APOBEC3B. IMPORTANCE Human cytomegalovirus (HCMV) infections can range from asymptomatic to severe, particularly in neonates and immunocompromised patients. HCMV has evolved strategies to overcome host-encoded antiviral defenses to achieve lytic viral DNA replication and dissemination and, under some conditions, latency and long-term persistence. Here, we show that HCMV infection causes the antiviral factor, APOBEC3B, to relocalize from the nuclear compartment to the cytoplasm. This overall strategy resembles that used by related herpesviruses. However, the HCMV relocalization mechanism utilizes a different viral factor(s) and available evidence suggests the involvement of at least one protein expressed at the early stages of infection. This knowledge is important because a greater understanding of this mechanism could lead to novel antiviral strategies that enable APOBEC3B to naturally restrict HCMV infection.
Keywords: APOBEC3B (A3B); herpesviruses; human cytomegalovirus; immediate-early genes; innate immunity; ribonucleotide reductase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Update of
-
Human cytomegalovirus mediates APOBEC3B relocalization early during infection through a ribonucleotide reductase-independent mechanism.bioRxiv [Preprint]. 2023 Jan 31:2023.01.30.526383. doi: 10.1101/2023.01.30.526383. bioRxiv. 2023. Update in: J Virol. 2023 Aug 31;97(8):e0078123. doi: 10.1128/jvi.00781-23 PMID: 36778493 Free PMC article. Updated. Preprint.
Similar articles
-
Human cytomegalovirus mediates APOBEC3B relocalization early during infection through a ribonucleotide reductase-independent mechanism.bioRxiv [Preprint]. 2023 Jan 31:2023.01.30.526383. doi: 10.1101/2023.01.30.526383. bioRxiv. 2023. Update in: J Virol. 2023 Aug 31;97(8):e0078123. doi: 10.1128/jvi.00781-23 PMID: 36778493 Free PMC article. Updated. Preprint.
-
A Conserved Mechanism of APOBEC3 Relocalization by Herpesviral Ribonucleotide Reductase Large Subunits.J Virol. 2019 Nov 13;93(23):e01539-19. doi: 10.1128/JVI.01539-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534038 Free PMC article.
-
Evidence linking APOBEC3B genesis and evolution of innate immune antagonism by gamma-herpesvirus ribonucleotide reductases.Elife. 2022 Dec 2;11:e83893. doi: 10.7554/eLife.83893. Elife. 2022. PMID: 36458685 Free PMC article.
-
APOBECs and Herpesviruses.Viruses. 2021 Feb 28;13(3):390. doi: 10.3390/v13030390. Viruses. 2021. PMID: 33671095 Free PMC article. Review.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
Cited by
-
Unveiling the Connection: Viral Infections and Genes in dNTP Metabolism.Viruses. 2024 Sep 3;16(9):1412. doi: 10.3390/v16091412. Viruses. 2024. PMID: 39339888 Free PMC article. Review.
-
Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes.bioRxiv [Preprint]. 2023 Jun 28:2023.06.27.546779. doi: 10.1101/2023.06.27.546779. bioRxiv. 2023. Update in: Virus Evol. 2023 Aug 02;9(2):vead047. doi: 10.1093/ve/vead047 PMID: 37425914 Free PMC article. Updated. Preprint.
-
Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes.Virus Evol. 2023 Aug 2;9(2):vead047. doi: 10.1093/ve/vead047. eCollection 2023. Virus Evol. 2023. PMID: 37577211 Free PMC article.
References
-
- Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. 2011. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–375. doi:10.1038/nature10693 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

