A ligand-receptor interactome atlas of the zebrafish
- PMID: 37539027
- PMCID: PMC10393773
- DOI: 10.1016/j.isci.2023.107309
A ligand-receptor interactome atlas of the zebrafish
Abstract
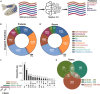
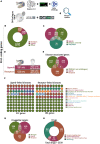
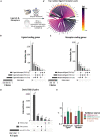
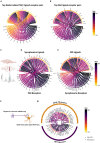
Studies in zebrafish can unravel the functions of cellular communication and thus identify novel bench-to-bedside drugs targeting cellular communication signaling molecules. Due to the incomplete annotation of zebrafish proteome, the knowledge of zebrafish receptors, ligands, and tools to explore their interactome is limited. To address this gap, we de novo predicted the cellular localization of zebrafish reference proteome using deep learning algorithm. We combined the predicted and existing annotations on cellular localization of zebrafish proteins and created repositories of zebrafish ligands, membrane receptome, and interactome as well as associated diseases and targeting drugs. Unlike other tools, our interactome atlas is based on both the physical interaction data of zebrafish proteome and existing human ligand-receptor pair databases. The resources are available as R and Python scripts. DanioTalk provides a novel resource for researchers interested in targeting cellular communication in zebrafish, as we demonstrate in applications studying synapse and axo-glial interactome. DanioTalk methodology can be applied to build and explore the ligand-receptor atlas of other non-mammalian model organisms.
Keywords: Cell biology; Omics; Specialized functions of cells.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Mycobacterium tuberculosis and Clostridium difficille interactomes: demonstration of rapid development of computational system for bacterial interactome prediction.Microb Inform Exp. 2012 Mar 21;2:4. doi: 10.1186/2042-5783-2-4. Microb Inform Exp. 2012. PMID: 22587966 Free PMC article.
-
Matching receptome genes with their ligands for surveying paracrine/autocrine signaling systems.Mol Endocrinol. 2007 Aug;21(8):2009-14. doi: 10.1210/me.2007-0087. Epub 2007 Jun 5. Mol Endocrinol. 2007. PMID: 17550980 Review.
-
Quantitative proteomics reveals CLR interactome in primary human cells.J Biol Chem. 2024 Jun;300(6):107399. doi: 10.1016/j.jbc.2024.107399. Epub 2024 May 20. J Biol Chem. 2024. PMID: 38777147 Free PMC article.
-
Integrated intra- and intercellular signaling knowledge for multicellular omics analysis.Mol Syst Biol. 2021 Mar;17(3):e9923. doi: 10.15252/msb.20209923. Mol Syst Biol. 2021. PMID: 33749993 Free PMC article.
-
Incorporating zebrafish omics into chemical biology and toxicology.Zebrafish. 2010 Mar;7(1):41-52. doi: 10.1089/zeb.2009.0636. Zebrafish. 2010. PMID: 20384484 Review.
Cited by
-
scRNA-seq reveals the diversity of the developing cardiac cell lineage and molecular players in heart rhythm regulation.iScience. 2024 May 22;27(6):110083. doi: 10.1016/j.isci.2024.110083. eCollection 2024 Jun 21. iScience. 2024. PMID: 38872974 Free PMC article.
-
Targeting autophagy impairment improves the phenotype of a novel CLN8 zebrafish model.Neurobiol Dis. 2024 Jul;197:106536. doi: 10.1016/j.nbd.2024.106536. Epub 2024 May 17. Neurobiol Dis. 2024. PMID: 38763444 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

