Development of a DNA aptamer targeting IDO1 with anti-tumor effects
- PMID: 37520707
- PMCID: PMC10374466
- DOI: 10.1016/j.isci.2023.107367
Development of a DNA aptamer targeting IDO1 with anti-tumor effects
Abstract
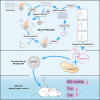
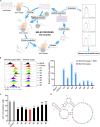
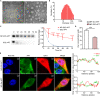
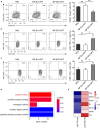
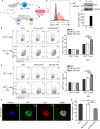
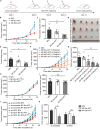
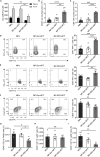
Immune checkpoint blockade has become an effective approach to reverse the immune tolerance of tumor cells. Indoleamine 2,3-dioxygenase 1 (IDO1) is frequently upregulated in many types of cancers and contributes to the establishment of an immunosuppressive cancer microenvironment, which has been thought to be a potential target for cancer therapy. However, the development of IDO1 inhibitors for clinical application is still limited. Here, we isolated a DNA aptamer with a strong affinity and inhibitory activity against IDO1, designated as IDO-APT. By conjugating with nanoparticles, in situ injection of IDO-APT to CT26 tumor-bearing mice significantly suppresses the activity of regulatory T cells and promotes the function of CD8+ T cells, leading to tumor suppression and prolonged survival. Therefore, this functional IDO1-specific aptamer with potent anti-tumor effects may serve as a potential therapeutic strategy in cancer immunotherapy. Our data provide an alternative way to target IDO1 in addition to small molecule inhibitors.
Keywords: Cancer; Immunology; Pharmacology.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Peptide vaccination directed against IDO1-expressing immune cells elicits CD8+ and CD4+ T-cell-mediated antitumor immunity and enhanced anti-PD1 responses.J Immunother Cancer. 2020 Jul;8(2):e000605. doi: 10.1136/jitc-2020-000605. J Immunother Cancer. 2020. PMID: 32690770 Free PMC article.
-
Aptamer-conjugated nano-liposome for immunogenic chemotherapy with reversal of immunosuppression.J Control Release. 2022 Aug;348:893-910. doi: 10.1016/j.jconrel.2022.06.039. Epub 2022 Jun 29. J Control Release. 2022. PMID: 35760233
-
Carboxyamidotriazole combined with IDO1-Kyn-AhR pathway inhibitors profoundly enhances cancer immunotherapy.J Immunother Cancer. 2019 Sep 11;7(1):246. doi: 10.1186/s40425-019-0725-7. J Immunother Cancer. 2019. PMID: 31511064 Free PMC article.
-
Indoleamine 2,3-dioxygenase 1 in circumventing checkpoint inhibitor responses: Updated.Int Immunopharmacol. 2023 May;118:110032. doi: 10.1016/j.intimp.2023.110032. Epub 2023 Mar 16. Int Immunopharmacol. 2023. PMID: 36933494 Review.
-
Updates in the Clinical Development of Epacadostat and Other Indoleamine 2,3-Dioxygenase 1 Inhibitors (IDO1) for Human Cancers.Front Oncol. 2018 Oct 4;8:423. doi: 10.3389/fonc.2018.00423. eCollection 2018. Front Oncol. 2018. PMID: 30338242 Free PMC article. Review.
References
-
- Takikawa O., Yoshida R., Kido R., Hayaishi O. Tryptophan degradation in mice initiated by indoleamine 2,3-dioxygenase. J. Biol. Chem. 1986;261:3648–3653. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

