Pleiotropy drives evolutionary repair of the responsiveness of polarized cell growth to environmental cues
- PMID: 37520345
- PMCID: PMC10382278
- DOI: 10.3389/fmicb.2023.1076570
Pleiotropy drives evolutionary repair of the responsiveness of polarized cell growth to environmental cues
Abstract
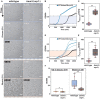
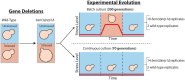
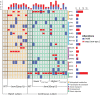
The ability of cells to translate different extracellular cues into different intracellular responses is vital for their survival in unpredictable environments. In Saccharomyces cerevisiae, cell polarity is modulated in response to environmental signals which allows cells to adopt varying morphologies in different external conditions. The responsiveness of cell polarity to extracellular cues depends on the integration of the molecular network that regulates polarity establishment with networks that signal environmental changes. The coupling of molecular networks often leads to pleiotropic interactions that can make it difficult to determine whether the ability to respond to external signals emerges as an evolutionary response to environmental challenges or as a result of pleiotropic interactions between traits. Here, we study how the propensity of the polarity network of S. cerevisiae to evolve toward a state that is responsive to extracellular cues depends on the complexity of the environment. We show that the deletion of two genes, BEM3 and NRP1, disrupts the ability of the polarity network to respond to cues that signal the onset of the diauxic shift. By combining experimental evolution with whole-genome sequencing, we find that the restoration of the responsiveness to these cues correlates with mutations in genes involved in the sphingolipid synthesis pathway and that these mutations frequently settle in evolving populations irrespective of the complexity of the selective environment. We conclude that pleiotropic interactions make a significant contribution to the evolution of networks that are responsive to extracellular cues.
Keywords: adaptation; cell architecture; cell polarity; fluctuating environment; laboratory evolution; phenotypic plasticity.
Copyright © 2023 Kingma, Diepeveen, Iñigo de la Cruz and Laan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Evolution of evolvability and phenotypic plasticity in virtual cells.BMC Evol Biol. 2017 Feb 28;17(1):60. doi: 10.1186/s12862-017-0918-y. BMC Evol Biol. 2017. PMID: 28241744 Free PMC article.
-
[Genetics and evolution of developmental plasticity in the nematode C. elegans: Environmental induction of the dauer stage].Biol Aujourdhui. 2020;214(1-2):45-53. doi: 10.1051/jbio/2020006. Epub 2020 Aug 10. Biol Aujourdhui. 2020. PMID: 32773029 Review. French.
-
The evolutionary plasticity of chromosome metabolism allows adaptation to constitutive DNA replication stress.Elife. 2020 Feb 11;9:e51963. doi: 10.7554/eLife.51963. Elife. 2020. PMID: 32043971 Free PMC article.
-
How to fit in: The learning principles of cell differentiation.PLoS Comput Biol. 2020 Apr 13;16(4):e1006811. doi: 10.1371/journal.pcbi.1006811. eCollection 2020 Apr. PLoS Comput Biol. 2020. PMID: 32282832 Free PMC article.
-
Pleiotropy in developmental regulation by flowering-pathway genes: is it an evolutionary constraint?New Phytol. 2019 Oct;224(1):55-70. doi: 10.1111/nph.15901. Epub 2019 Jun 18. New Phytol. 2019. PMID: 31074008 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

